Title
题目
TransUNet: Rethinking the U-Net architecture design for medical imagesegmentation through the lens of transformers
TransUNet: 通过Transformer的视角重新思考U-Net架构在医学图像分割中的设计
01
文献速递介绍
卷积神经网络(CNNs),特别是全卷积网络(FCNs)(Long 等,2015),在医学图像分割领域中获得了显著的关注。在其各种迭代模型中,U-Net 模型(Ronneberger 等,2015)因其对称的编码器–解码器设计,并通过跳跃连接增强细节保留,成为许多研究人员的首选。基于这一方法,各类医学成像任务中取得了显著进展。这些进展包括磁共振成像(MRI)中的心脏分割(Yu 等,2017)、利用计算机断层扫描(CT)进行的器官勾勒(Zhou 等,2017;Li 等,2018b;Yu 等,2018;Luo 等,2021)以及结肠镜检查中的息肉分割(Zhou 等,2019)。
尽管CNN在表示能力方面无可匹敌,但由于卷积操作的局部性,在建模远程关系时往往表现不足。当面对不同患者之间纹理、形状和大小的巨大变化时,这一局限性尤其明显。认识到这一不足,研究界越来越倾向于使用完全基于注意力机制的Transformers模型,因为它们在捕捉全局上下文方面有着天然的优势(Vaswani 等,2017)。然而,Transformers将输入处理为一维序列,优先进行全局上下文建模,容易生成低分辨率的特征。因此,一种更有前景的混合方法是结合CNN和Transformer编码器。
TransUNet(Chen 等,2021)于2021年首次提出,是首批将Transformer集成到医学图像分析中的模型之一。该方法利用了U-Net编码器的高分辨率空间细节,同时发挥了Transformers在全局上下文建模中的优势,这在医学图像分割中至关重要。这一创新促使了后续研究的开展(Cao 等,2022;Xie 等,2021;Hatamizadeh 等,2021)。尽管如此,不同U-Net组件中Transformers自注意力机制的全面理解仍然缺失。
Abatract
摘要
Medical image segmentation is crucial for healthcare, yet convolution-based methods like U-Net face limitationsin modeling long-range dependencies. To address this, Transformers designed for sequence-to-sequencepredictions have been integrated into medical image segmentation. However, a comprehensive understandingof Transformers’ self-attention in U-Net components is lacking. TransUNet, first introduced in 2021, is widelyrecognized as one of the first models to integrate Transformer into medical image analysis. In this study,we present the versatile framework of TransUNet that encapsulates Transformers’ self-attention into two keymodules: (1) a Transformer encoder tokenizing image patches from a convolution neural network (CNN)feature map, facilitating global context extraction, and (2) a Transformer decoder refining candidate regionsthrough cross-attention between proposals and U-Net features. These modules can be flexibly inserted intothe U-Net backbone, resulting in three configurations: Encoder-only, Decoder-only, and Encoder+Decoder.TransUNet provides a library encompassing both 2D and 3D implementations, enabling users to easily tailorthe chosen architecture. Our findings highlight the encoder’s efficacy in modeling interactions among multipleabdominal organs and the decoder’s strength in handling small targets like tumors. It excels in diversemedical applications, such as multi-organ segmentation, pancreatic tumor segmentation, and hepatic vesselsegmentation. Notably, our TransUNet achieves a significant average Dice improvement of 1.06% and 4.30%for multi-organ segmentation and pancreatic tumor segmentation, respectively, when compared to the highlycompetitive nn-UNet, and surpasses the top-1 solution in the BrasTS2021 challenge.
医学图像分割在医疗保健中至关重要,但基于卷积的U-Net方法在建模远程依赖关系方面存在局限性。为了解决这一问题,Transformer被设计用于序列到序列的预测,并被集成到医学图像分割中。然而,目前对Transformer自注意力机制在U-Net组件中的全面理解仍然缺乏。TransUNet首次于2021年提出,被广泛认为是将Transformer集成到医学图像分析中的首批模型之一。
在本研究中,我们展示了TransUNet的通用框架,该框架将Transformer的自注意力机制封装到两个关键模块中:(1) Transformer编码器将卷积神经网络(CNN)特征图中的图像块进行标记,有助于提取全局上下文;(2) Transformer解码器通过提案与U-Net特征之间的交叉注意力机制,优化候选区域。这些模块可以灵活地插入到U-Net的主干结构中,形成三种配置:仅编码器、仅解码器、编码器+解码器。
TransUNet提供了包含2D和3D实现的库,用户可以轻松定制所选架构。我们的研究结果表明,编码器在建模多个腹部器官之间的交互方面表现优异,而解码器在处理肿瘤等小目标时表现突出。它在多种医学应用中表现出色,如多器官分割、胰腺肿瘤分割和肝脏血管分割。值得注意的是,与竞争力很强的nn-UNet相比,TransUNet在多器官分割和胰腺肿瘤分割中分别实现了1.06%和4.30%的Dice平均提升,并超越了BrasTS2021挑战赛的排名第一的解决方案。
Method
方法
Given a 3D medical image (e.g., CT/MR scan) 𝐱 ∈ R𝐷×𝐻×𝑊 ×𝐶 withthe spatial resolution of 𝐷×𝐻×𝑊 and 𝐶* number of channels. We aim topredict the corresponding pixel-wise labelmap with size 𝐷×𝐻 ×𝑊 . Themost common way is to directly train a CNN (e.g., U-Net) to first encodeimages into high-level feature representations, which are then decodedback to the full spatial resolution. Our approach diverges from conventional methods by thoroughly exploring the attention mechanismsutilized in both the encoder and decoder phases of standard U-shapedsegmentation architectures, employing Transformers. In Section 3.1, wedelve into the direct application of Transformers for encoding featurerepresentations from segmented image patches. Following this, in Section 3.2, we elaborate on implementing the query-based Transformer,which serves as our decoder. The detailed architecture of TransUNet isthen presented in Section 3.3.
给定一个3D医学图像(如CT/MR扫描)𝐱 ∈ R𝐷×𝐻×𝑊 ×𝐶,其空间分辨率为𝐷×𝐻×𝑊,并且包含𝐶个通道。我们的目标是预测相应的像素级标签图,大小为𝐷×𝐻×𝑊*。最常见的方法是直接训练一个卷积神经网络(CNN,例如U-Net),首先将图像编码为高层次的特征表示,然后解码回完整的空间分辨率。我们的方法不同于传统方法,深入探索了在标准U形分割架构中编码器和解码器阶段使用的注意力机制,并采用了Transformers。
在第3.1节中,我们探讨了直接应用Transformers对分割后的图像块进行特征表示编码。接下来,在第3.2节中,我们详细介绍了基于查询的Transformer的实现,它作为我们的解码器。TransUNet的详细架构则在第3.3节中进行呈现。
Conclusion
结论
While U-Net has been successful, its limitations in handling longrange dependencies have prompted the exploration of Transformer asan alternative architecture. In this work, we introduce a Transformercentric encoder–decoder framework, named TransUNet. Specifically,we introduce (1) A Transformer encoder that tokenizes CNN featuremap patches, facilitating a richer extraction of global contexts; and(2) A Transformer decoder designed to adaptively refine segmentationregions, capitalizing on cross-attention mechanisms between candidateproposals and U-Net features. We also propose a coarse-to-fine attentionrefinement to enhance the segmentation of small targets and tumorsin the Transformer decoder. Through extensive experimentation, weprovide the first thorough investigation on the impact of integrating theTransformer encoder and decoder into U-Net architectures, providinginsights for addressing diverse challenges in medical image segmentation. Empirical results showcase TransUNet’s superior performancein multi-organ, pancreatic tumor, hepatic vessel and tumor segmentation. Additionally, we surpassed top-1 solution in the BraTS2021challenge. Additionally, we have released our codebase to facilitatefurther exploration and encourage the adoption of Transformers inmedical applications, offering both 2D and 3D implementations for userconvenience.
尽管U-Net在医学图像分割中取得了成功,但其在处理远距离依赖关系上的局限性促使人们探索Transformer作为替代架构。在这项工作中,我们引入了一个以Transformer为核心的编码器-解码器框架,称为TransUNet。具体而言,我们引入了:(1) 一个Transformer编码器,用于对CNN特征图块进行标记,促进更丰富的全局上下文提取;(2) 一个Transformer解码器,设计用于自适应地细化分割区域,利用候选提议和U-Net特征之间的交叉注意力机制。我们还提出了一种从粗到精的注意力细化策略,以增强Transformer解码器中对小目标和肿瘤的分割。通过广泛的实验,我们首次对将Transformer编码器和解码器集成到U-Net架构中的影响进行了深入研究,为解决医学图像分割中的多样化挑战提供了见解。实验证明,TransUNet在多器官、胰腺肿瘤、肝血管和肿瘤分割中表现优异。此外,我们超越了BraTS2021挑战赛中的top-1解决方案。我们还发布了代码库,便于进一步探索和促进Transformer在医学应用中的采用,提供了2D和3D实现以方便用户使用。
Figure
图

Fig. 1. Overview of TransUNet. Our proposed architecture consists of two components: (1) the Transformer encoder where a CNN encoder is firstly used for local image featureextraction, followed by a pure Transformer encoder for global information interaction; and (2) the Transformer decoder that reframes per-pixel segmentation as mask classificationusing learnable queries, which are refined through cross-attention with CNN features, and employs a coarse-to-fine attention refinement approach for enhanced segmentationaccuracy.
图 1. TransUNet 概览。我们提出的架构由两个组件组成:(1) Transformer 编码器,其中首先使用 CNN 编码器进行局部图像特征提取,随后通过纯 Transformer 编码器进行全局信息交互;(2) Transformer 解码器,将每像素分割重新构建为基于可学习查询的掩码分类,通过与 CNN 特征的交叉注意力机制进行细化,并采用粗到细的注意力优化方法以提高分割精度。
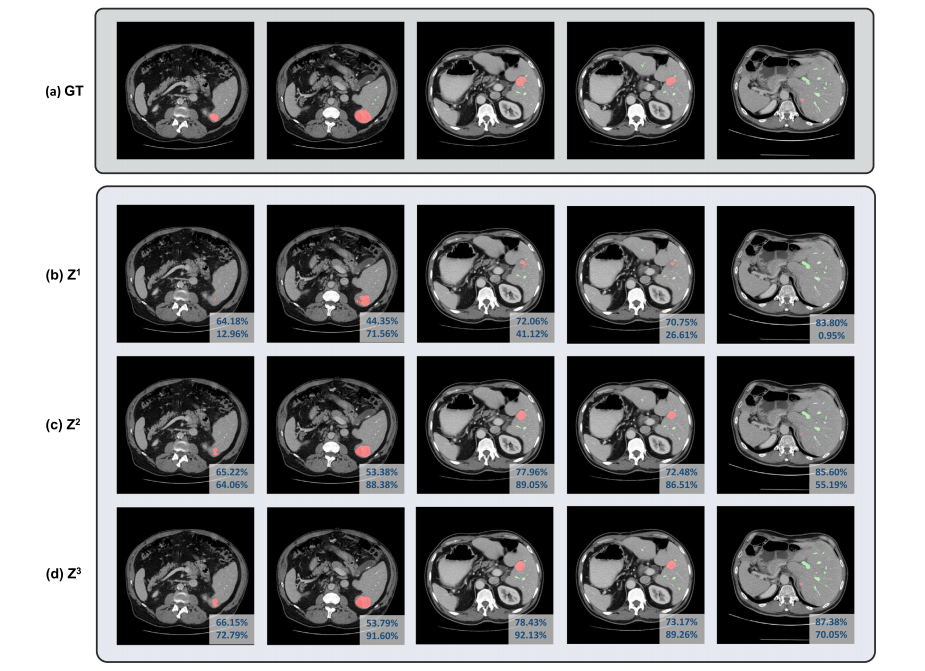
Fig. 2. Visualizations of outputs from different iterations during coarse-to-fine refinement: (a) Groundtruth. (b-d) the segmentation mask at the first to the third iteration. Differentcolumns represent different samples from MSD Vessel Dataset. The dice coefficients of vessels and tumors are indicated in each image’s first and second row of the lower rightcorner, respectively.
图2. 不同迭代过程中从粗到精细优化的输出可视化:(a) 真实标注。(b-d) 第一到第三次迭代的分割掩膜。不同列表示来自MSD血管数据集的不同样本。每张图像右下角第一行和第二行分别标明了血管和肿瘤的Dice系数。
Table
表
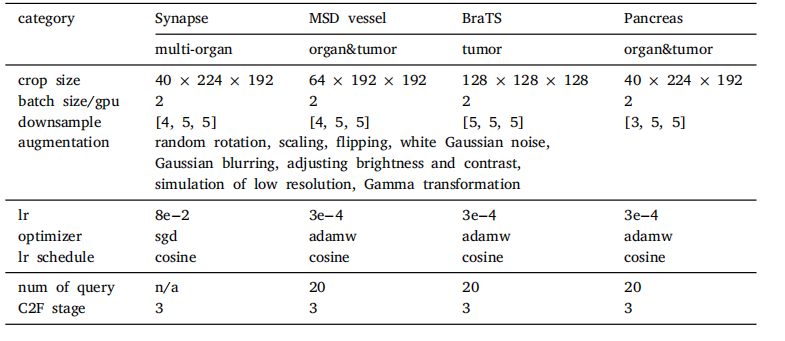
Table 1Implementation details including the architecture hyperparameters, training settings,and data augmentation. Note that the customized hyperparameters (row3-6) are directlyborrowed from nnUNet configuration.
表 1 实现细节,包括架构超参数、训练设置和数据增强。请注意,定制的超参数(第3-6行)直接借用了nnUNet的配置。

Table 2Comparison of different configurations of TransUNet on the BTCV multi-organ CT dataset (average dice score %, and dice score % for each organ).
表 2 不同配置的 TransUNet 在 BTCV 多器官 CT 数据集上的比较(平均 Dice 得分百分比,以及每个器官的 Dice 得分百分比)。

Table 3Comparison of different configurations of TransUNet on MSD vessel dataset with dicescore metrics (%). Experiments are conducted in five-fold cross-validation
表 3 不同配置的 TransUNet 在 MSD 血管数据集上的比较,使用 Dice 得分指标(百分比)。实验在五折交叉验证中进行。
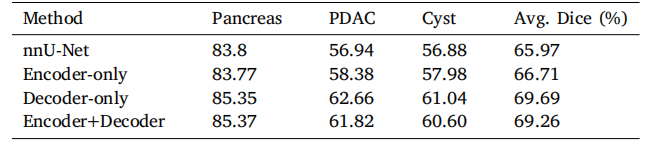
Table 4Generalization of the Transformer decoder to different pancreatic tumors on ourin-house large-scale pancreatic tumor segmentation dataset.
表 4 Transformer 解码器在不同胰腺肿瘤上的泛化能力测试,基于我们内部的大规模胰腺肿瘤分割数据集。
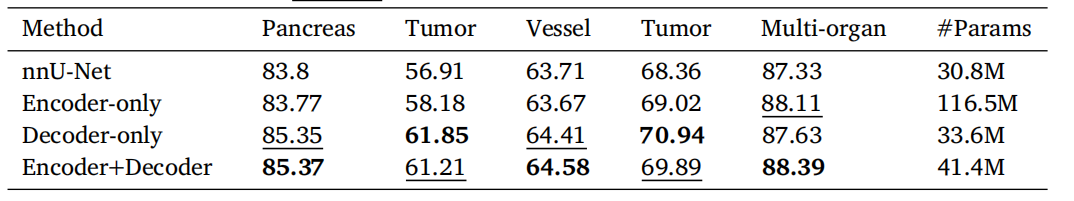
Table 5Performance (average Dice (%)) v.s. network parameters comparison on MSD HepaticVessel dataset, ourin-house large-scale pancreatic mass dataset, and Synapse multi-organ segmentation dataset. The pancreatictumor Dice scores are averaged from the performance of both pancreatic cyst and PDAC segmentation. bolddenotes the best results and underline denotes the second best results.
表 5 MSD 肝血管数据集、我们内部的大规模胰腺肿块数据集以及 Synapse 多器官分割数据集上的性能(平均 Dice 得分百分比)与网络参数的比较。胰腺肿瘤的 Dice 得分是胰腺囊肿和胰腺导管腺癌(PDAC)分割性能的平均值。粗体表示最佳结果,下划线表示次优结果。
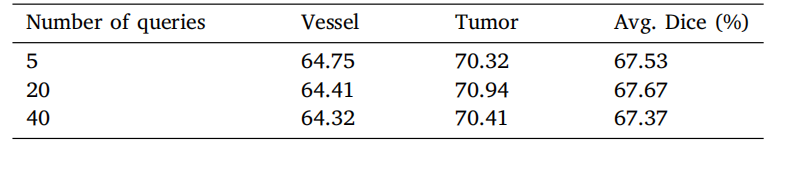
Table 6Ablation of number of queries under Transformer decoder setting on MSD vessel datasetwith dice score metrics (%). Experiments are conducted in five-fold cross-validation.
表6 Transformer解码器设置下关于查询数量消融实验的结果(基于MSD血管数据集的Dice分数指标,单位:%)。实验在五折交叉验证中进行。
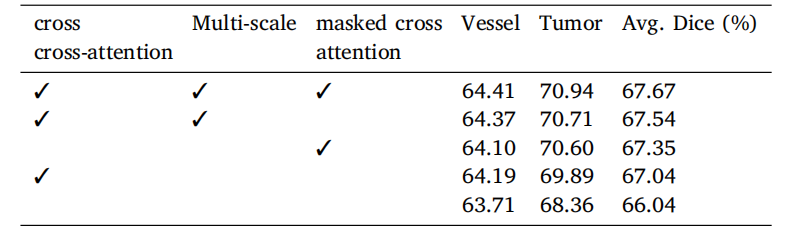
Table 7Ablation of different types of attention mechanisms for the Transformer decoder onMSD vessel dataset with dice score metrics (%). Experiments are conducted in five-foldcross-validation.
表7 在MSD血管数据集上,不同类型的注意力机制在Transformer解码器中的消融实验结果(基于Dice分数指标,单位:%)。实验在五折交叉验证中进行。
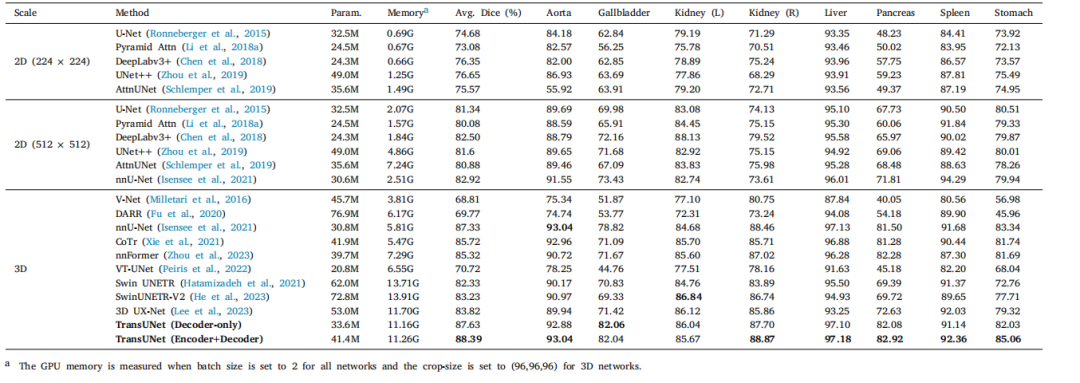
Table 8Comparison on the BTCV multi-organ CT dataset (average dice score %, and dice score % for each organ).
表8 BTCV多器官CT数据集上的对比结果(平均Dice分数%,以及每个器官的Dice分数%)。
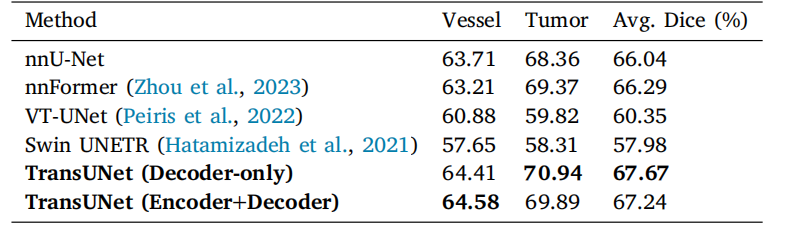
Table 9Performance comparison on MSD vessel dataset with dice score metrics (%).Experiments are conducted in five-fold cross-validation.
表9 MSD血管数据集上性能对比结果(基于Dice分数指标,单位:%)。实验在五折交叉验证中进行。
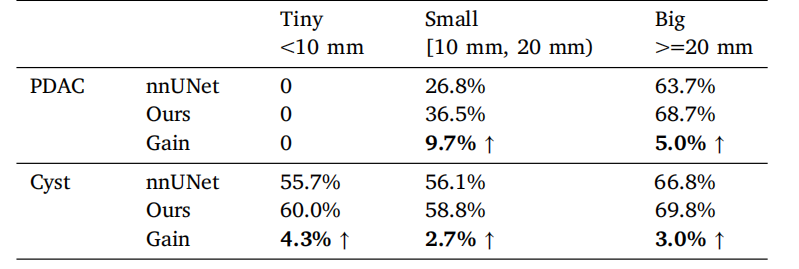
Table 10Tumor segmentation performance (average Dice) under different tumor sizes reportedin the pancreatic tumor dataset.
表10 胰腺肿瘤数据集中,不同肿瘤大小下的肿瘤分割性能(平均Dice分数)。
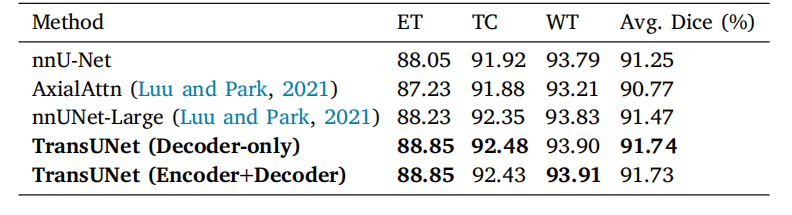
Table 11Performance comparison on the BraTS2021 challenge for brain tumor segmentationwith dice score metrics (%). Experiments are conducted in five-fold cross-validation.
表11 BraTS2021挑战赛中的脑肿瘤分割性能对比结果(基于Dice分数指标,单位:%)。实验在五折交叉验证中进行。
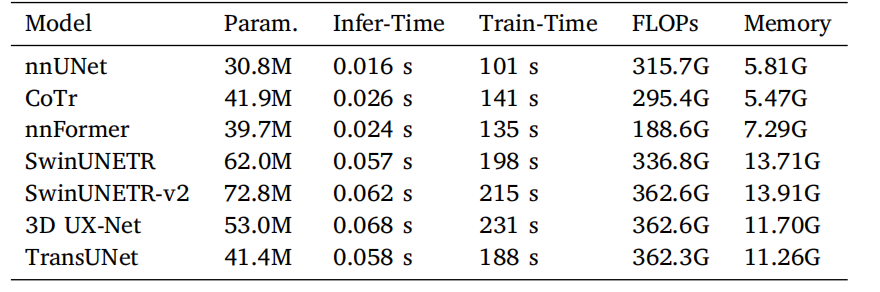
Table 12Comparison of different 3D models based on inference time (seconds per volume),training time (seconds per epoch), FLOPs, and GPU memory usage.
表12 不同3D模型在推理时间(每体积的秒数)、训练时间(每轮的秒数)、FLOPs(浮点运算次数)以及GPU内存使用方面的对比。



















