1.安装 htslib-1.20
https://www.htslib.org/doc/tabix.html
@J3$ cd ~/Downloads/
$ wget https://github.com/samtools/htslib/releases/download/1.20/htslib-1.20.tar.bz2
$ tar jxvf htslib-1.20.tar.bz2
编译安装:
$ cd htslib-1.20/
$ ./configure --prefix=/home/wangjl/soft/htslib-1.20/
$ make -j16
$ make install
配置路径:
$ vim ~/.bashrc
export PATH=$PATH:/home/wangjl/soft/htslib-1.20/bin
测试:
$ source ~/.bashrc
$ which bgzip
~/soft/htslib-1.20/bin/bgzip
$ which tabix
~/soft/htslib-1.20/bin/tabix
2. 生成fa和gtf的索引文件fa.fai, gtf.gz.tbi
$ cd /data/wangjl/scPolyA-seq2/ref/hg38/gencode/
$ (grep ^"#" GRCh38.p13.gtf; grep -v ^"#" GRCh38.p13.gtf | sort -t $'\t' -k1,1V -k4,4n -k5,5n) | bgzip > GRCh38.p13.sorted.gtf.gz
$ tabix -p gff GRCh38.p13.sorted.gtf.gz
$ samtools faidx input_ref.fa #生成 input_ref.fa.fai
$ ls -lth
-rw-r--r--. 1 wangjl jinlab 370K Jul 24 20:03 GRCh38.p13.sorted.gtf.gz.tbi
-rw-r--r--. 1 wangjl jinlab 36M Jul 24 20:00 GRCh38.p13.sorted.gtf.gz
-rw-r--r--. 1 wangjl jinlab 905M May 1 2023 GRCh38.p13.gtf
-rw-r--r--. 1 wangjl jinlab 22K May 8 2023 GRCh38.p13.genome.fa.fai
-rw-r--r--. 1 wangjl jinlab 3.1G May 1 2023 GRCh38.p13.genome.fa
3.在IGV.js中引用资源
其他资源使用默认的,我已下载到服务器本地。
// refer in local: gencode
var hg38_local_gencode={
"id": "hg38",
"name": "Human (GRCh38/hg38)",
"fastaURL": "ref/hg38/gencode/GRCh38.p13.genome.fa", //?someRandomSeed=0
"indexURL": "ref/hg38/gencode/GRCh38.p13.genome.fa.fai",
"cytobandURL": "ref/hg38/cytoBandIdeo.txt.gz",
"aliasURL": "ref/hg38/hg38_alias.tab",
"tracks": [
{
"name": "GRCh38",
"format": "gtf",
"type": "annotation",
"id": "hg38_genes",
"url": "ref/hg38/gencode/GRCh38.p13.sorted.gtf.gz",
"indexURL": "ref/hg38/gencode/GRCh38.p13.sorted.gtf.gz.tbi",
"visibilityWindow": -1,
//"visibilityWindow": 10000000, // 轨道的可见窗口大小
"supportsWholeGenome": false,
"removable": false,
"order": 1000000,
"height":250, //height of ref track
"infoURL": "https://www.ncbi.nlm.nih.gov/gene/?term=$$" //links to ncbi gene
}
],
"chromosomeOrder": "chr1, chr2, chr3, chr4, chr5, chr6, chr7, chr8, chr9, chr10, chr11, chr12, chr13, chr14, chr15, chr16, chr17, chr18, chr19, chr20, chr21, chr22, chrX, chrY"
}
var options =
{
//genome: "hg38",
//init screen
"locus": "chr21:45,512,381-45,521,866",
reference:hg38_local_gencode,
tracks: []
};
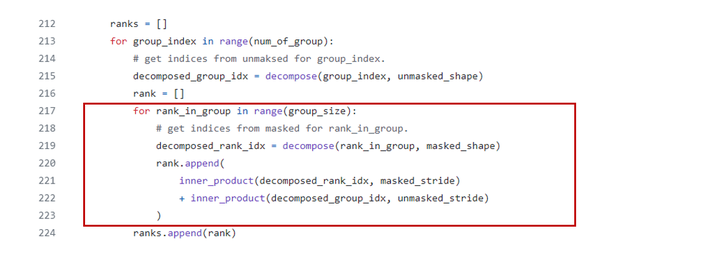
测试结果
- 上图:IGV默认的基因组注释文件
ref/hg38/ncbiRefSeq.txt.gz - 下图:我们导入的基因组注释文件
ref/hg38/gencode/GRCh38.p13.sorted.gtf.gz
两者在同一个地方还是有一些差异的:下图甚至多出一个转录本。

不过,不能评价哪个注释更可靠。毕竟都是知名学术机构发布的。
建议在一个项目中,使用最新的基因组注释文件,且
- 固定使用同一个机构、同一个版本号的gtf文件,
- 保证gtf和fa配对。