本文无文本解析,结尾处有简单装箱问题的示例,该算法收敛结果较慢,仅供ABAP爱好者参考,实践,实际应用建议使用线性规划。可直接复制后在系统中使用。
对象自定义逻辑版本-截图
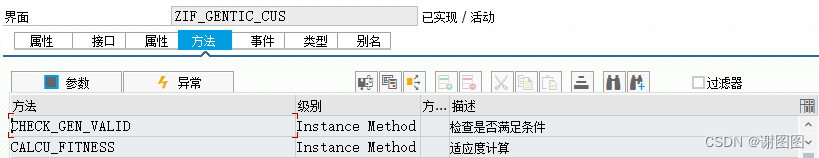
对象自定义逻辑版本-对象描述
INIT Instance Method Public 初始化
ADD_VAR Instance Method Public 添加变量
GET_RESULT Instance Method Public 获取最佳结果
GET_RECORD Instance Method Public 获取过程记录
CALCU_FITNESS Instance Method Protected 适应度计算
ELIMINATE Instance Method Protected 淘汰个体
GEN_INI_ETHNICGROUPS Instance Method Private 生成初始化族群
CALCULAT_RATE Instance Method Private 计算概率
UNIT_VAR Instance Method Private 变异
UNIT_CROSS Instance Method Private 交叉
RANDOW Instance Method Private 随机数
CHECK_GEN_VALID Instance Method Private 检查是否满足条件
GEN_UNIT Instance Method Private 生成单个染色体
ITERATION Instance Method Private 迭代繁衍群体
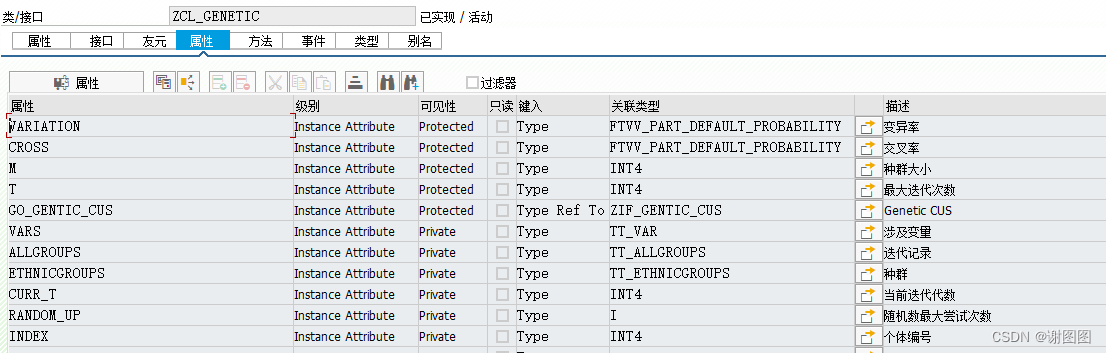
VARIATION Instance Attribute Protected Type FTVV_PART_DEFAULT_PROBABILITY 变异率
CROSS Instance Attribute Protected Type FTVV_PART_DEFAULT_PROBABILITY 交叉率
M Instance Attribute Protected Type INT4 种群大小
T Instance Attribute Protected Type INT4 最大迭代次数
GO_GENTIC_CUS Instance Attribute Protected Type Ref To ZIF_GENTIC_CUS Genetic CUS
VARS Instance Attribute Private Type TT_VAR 涉及变量
ALLGROUPS Instance Attribute Private Type TT_ALLGROUPS 迭代记录
ETHNICGROUPS Instance Attribute Private Type TT_ETHNICGROUPS 种群
CURR_T Instance Attribute Private Type INT4 当前迭代代数
RANDOM_UP Instance Attribute Private Type I 随机数最大尝试次数
INDEX Instance Attribute Private Type INT4 个体编号对象自定义逻辑版本-类源代码
class ZCL_GENETIC definition
public
create public .
public section.
types:
BEGIN OF ty_allgroups,
t TYPE i,
index TYPE i,
units TYPE chip_property_tab,
fitness TYPE ftvv_part_default_probability,
END OF ty_allgroups .
types:
tt_allgroups TYPE TABLE OF ty_allgroups .
methods INIT
importing
!IV_VARIATION type FTVV_PART_DEFAULT_PROBABILITY default '0.05'
!IV_CROSS type FTVV_PART_DEFAULT_PROBABILITY default '0.8'
!IV_M type INT4 default 50
!IV_T type INT4 default 50
!IV_RANDOM_UP type I default 100
!IO_GENETIC_CUS type ref to ZIF_GENTIC_CUS .
methods ADD_VAR
importing
!NAME type STRING
!LOW type I
!HIGH type I .
methods GET_RESULT
returning
value(RT_UNITS) type CHIP_PROPERTY_TAB .
methods GET_RECORD
exporting
value(ET_REC) type ZCL_GENETIC_BASE=>TT_ALLGROUPS .
protected section.
types:
BEGIN OF ty_var,
name TYPE string,
low TYPE i,
high TYPE i,
END OF ty_var .
types:
tt_var TYPE TABLE OF ty_var .
types:
BEGIN OF ty_ethnicgroups,
index TYPE i,
units TYPE chip_property_tab,
fitness TYPE ftvv_part_default_probability,
END OF ty_ethnicgroups .
types:
tt_ethnicgroups TYPE TABLE OF ty_ethnicgroups .
data VARIATION type FTVV_PART_DEFAULT_PROBABILITY . " 变异概率
data CROSS type FTVV_PART_DEFAULT_PROBABILITY . " 交叉概率
data M type INT4 . " 初始种群大小
data T type INT4 . " 最大迭代次数
data GO_GENTIC_CUS type ref to ZIF_GENTIC_CUS .
methods CALCU_FITNESS
importing
value(UNITS) type CHIP_PROPERTY_TAB
returning
value(FITNESS) type FTVV_PART_DEFAULT_PROBABILITY .
methods ELIMINATE .
private section.
data VARS type TT_VAR ." 染色体
data ALLGROUPS type TT_ALLGROUPS ." 所有迭代记录
data ETHNICGROUPS type TT_ETHNICGROUPS ."种群
data CURR_T type INT4 ." 当前迭代次数
data RANDOM_UP type I ." 随机数重试最大次数
data INDEX type INT4 . " 个体ID
methods GEN_INI_ETHNICGROUPS .
methods CALCULAT_RATE
returning
value(RATE) type PAPLI .
methods UNIT_VAR
changing
!UNITS type CHIP_PROPERTY_TAB .
methods UNIT_CROSS
importing
!UNITS_FA type CHIP_PROPERTY_TAB
!UNITS_MO type CHIP_PROPERTY_TAB
returning
value(UNITS) type CHIP_PROPERTY_TAB .
methods RANDOW
importing
!LOW type I
!HIGH type I
returning
value(VALUE) type I .
methods CHECK_GEN_VALID
importing
!UNITS type CHIP_PROPERTY_TAB
returning
value(RV_BOOL) type CHAR1 .
methods GEN_UNIT
returning
value(UNITS) type CHIP_PROPERTY_TAB .
methods ITERATION .
ENDCLASS.
CLASS ZCL_GENETIC IMPLEMENTATION.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Public Method ZCL_GENETIC->ADD_VAR
* +-------------------------------------------------------------------------------------------------+
* | [--->] NAME TYPE STRING
* | [--->] LOW TYPE I
* | [--->] HIGH TYPE I
* +--------------------------------------------------------------------------------------</SIGNATURE>
METHOD ADD_VAR.
APPEND VALUE #( name = to_upper( name ) low = low high = high ) TO vars.
ENDMETHOD.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Private Method ZCL_GENETIC->CALCULAT_RATE
* +-------------------------------------------------------------------------------------------------+
* | [<-()] RATE TYPE PAPLI
* +--------------------------------------------------------------------------------------</SIGNATURE>
method CALCULAT_RATE.
rate = randow( low = 1 high = 1000 ) / 1000.
endmethod.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Protected Method ZCL_GENETIC->CALCU_FITNESS
* +-------------------------------------------------------------------------------------------------+
* | [--->] UNITS TYPE CHIP_PROPERTY_TAB
* | [<-()] FITNESS TYPE FTVV_PART_DEFAULT_PROBABILITY
* +--------------------------------------------------------------------------------------</SIGNATURE>
METHOD CALCU_FITNESS.
go_gentic_cus->calcu_fitness( EXPORTING units = units CHANGING fitness = fitness ).
ENDMETHOD.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Private Method ZCL_GENETIC->CHECK_GEN_VALID
* +-------------------------------------------------------------------------------------------------+
* | [--->] UNITS TYPE CHIP_PROPERTY_TAB
* | [<-()] RV_BOOL TYPE CHAR1
* +--------------------------------------------------------------------------------------</SIGNATURE>
METHOD check_gen_valid.
rv_bool = abap_true.
go_gentic_cus->check_gen_valid( EXPORTING units = units CHANGING cv_bool = rv_bool ).
ENDMETHOD.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Protected Method ZCL_GENETIC->ELIMINATE
* +-------------------------------------------------------------------------------------------------+
* +--------------------------------------------------------------------------------------</SIGNATURE>
METHOD ELIMINATE.
DATA: lv_count TYPE i,
lv_o TYPE p LENGTH 15,
lv_rate TYPE ftvv_part_default_probability.
DATA: lt_ethnicgroups LIKE ethnicgroups,
ls_ethnicgroups LIKE LINE OF ethnicgroups.
DO random_up TIMES.
lt_ethnicgroups = ethnicgroups.
DESCRIBE TABLE lt_ethnicgroups LINES lv_count.
IF ( lv_count - m ) / m <= 1 / 10.
EXIT.
ENDIF.
lv_o = lv_count * lv_count + lv_count.
SORT lt_ethnicgroups BY fitness DESCENDING .
LOOP AT lt_ethnicgroups INTO ls_ethnicgroups.
lv_rate = sy-tabix * sy-tabix / lv_o.
IF lv_rate < ( 1 / 200 ).
lv_rate = ( 1 / 200 ).
ENDIF.
IF calculat_rate( ) <= lv_rate.
DELETE ethnicgroups WHERE index = ls_ethnicgroups-index.
ENDIF.
ENDLOOP.
ENDDO.
ENDMETHOD.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Private Method ZCL_GENETIC->GEN_INI_ETHNICGROUPS
* +-------------------------------------------------------------------------------------------------+
* +--------------------------------------------------------------------------------------</SIGNATURE>
METHOD GEN_INI_ETHNICGROUPS.
DATA: ls_ethnicgroup LIKE LINE OF ethnicgroups.
DO m * 2 TIMES.
ls_ethnicgroup-units = gen_unit( ).
IF ls_ethnicgroup-units IS NOT INITIAL.
ls_ethnicgroup-index = index.
index = index + 1.
APPEND ls_ethnicgroup TO ethnicgroups.
ENDIF.
IF lines( ethnicgroups ) >= m.
EXIT.
ENDIF.
ENDDO.
ENDMETHOD.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Private Method ZCL_GENETIC->GEN_UNIT
* +-------------------------------------------------------------------------------------------------+
* | [<-()] UNITS TYPE CHIP_PROPERTY_TAB
* +--------------------------------------------------------------------------------------</SIGNATURE>
METHOD GEN_UNIT.
DATA: ls_unit LIKE LINE OF units,
lt_unit LIKE units,
ls_var LIKE LINE OF vars.
DO random_up TIMES.
CLEAR lt_unit.
LOOP AT vars INTO ls_var.
ls_unit-name = ls_var-name.
ls_unit-value = randow( low = ls_var-low high = ls_var-high ).
APPEND ls_unit TO lt_unit.
ENDLOOP.
IF check_gen_valid( lt_unit ).
EXIT.
ENDIF.
CLEAR lt_unit.
ENDDO.
units = lt_unit.
ENDMETHOD.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Public Method ZCL_GENETIC->GET_RECORD
* +-------------------------------------------------------------------------------------------------+
* | [<---] ET_REC TYPE ZCL_GENETIC_BASE=>TT_ALLGROUPS
* +--------------------------------------------------------------------------------------</SIGNATURE>
method GET_RECORD.
et_rec = allgroups.
endmethod.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Public Method ZCL_GENETIC->GET_RESULT
* +-------------------------------------------------------------------------------------------------+
* | [<-()] RT_UNITS TYPE CHIP_PROPERTY_TAB
* +--------------------------------------------------------------------------------------</SIGNATURE>
METHOD GET_RESULT.
DATA: lt_ethnicgroups LIKE ethnicgroups,
lv_index TYPE i.
gen_ini_ethnicgroups( ).
DO t TIMES.
iteration( ).
eliminate( ).
" 判定是否收敛
lt_ethnicgroups = ethnicgroups.
SORT lt_ethnicgroups BY fitness DESCENDING.
lv_index = lines( lt_ethnicgroups ) / 2.
IF lv_index >= 1 AND lt_ethnicgroups[ 1 ]-fitness = lt_ethnicgroups[ lv_index ]-fitness.
EXIT.
ENDIF.
IF curr_t > t.
EXIT.
ENDIF.
ENDDO.
IF ethnicgroups IS NOT INITIAL.
SORT ethnicgroups BY fitness DESCENDING.
rt_units = ethnicgroups[ 1 ]-units.
ENDIF.
ENDMETHOD.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Public Method ZCL_GENETIC->INIT
* +-------------------------------------------------------------------------------------------------+
* | [--->] IV_VARIATION TYPE FTVV_PART_DEFAULT_PROBABILITY (default ='0.05')
* | [--->] IV_CROSS TYPE FTVV_PART_DEFAULT_PROBABILITY (default ='0.8')
* | [--->] IV_M TYPE INT4 (default =50)
* | [--->] IV_T TYPE INT4 (default =50)
* | [--->] IV_RANDOM_UP TYPE I (default =100)
* | [--->] IO_GENETIC_CUS TYPE REF TO ZIF_GENTIC_CUS
* +--------------------------------------------------------------------------------------</SIGNATURE>
METHOD INIT.
*----------------------------------------------------------------------*
* variation: 变异率 0.01-0.1之间,变异率高可能导致收敛过快
* cross: 交叉率 0.6-0.9之间,交叉率越高范围越广
* m: 种群大小,20-200之间
* t: 最大迭代次数
* random_up: 随机时的重试上限
*----------------------------------------------------------------------*
variation = iv_variation .
cross = iv_cross .
m = iv_m .
t = iv_t .
random_up = iv_random_up .
go_gentic_cus = io_genetic_cus.
ENDMETHOD.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Private Method ZCL_GENETIC->ITERATION
* +-------------------------------------------------------------------------------------------------+
* +--------------------------------------------------------------------------------------</SIGNATURE>
METHOD ITERATION.
DATA: lv_fitnessall TYPE p LENGTH 15 DECIMALS 5,
lv_rate TYPE umls1.
DATA: ls_ethnicgroups LIKE LINE OF ethnicgroups,
lt_enthicgroups LIKE ethnicgroups,
ls_allgroups LIKE LINE OF allgroups,
lt_allgroups LIKE allgroups,
lt_units_ch TYPE chip_property_tab,
lt_units_fa TYPE chip_property_tab,
lt_units_mo TYPE chip_property_tab.
LOOP AT ethnicgroups INTO ls_ethnicgroups.
IF ls_ethnicgroups-fitness IS INITIAL.
ls_ethnicgroups-fitness = calcu_fitness( ls_ethnicgroups-units ).
MODIFY ethnicgroups FROM ls_ethnicgroups.
ENDIF.
lv_fitnessall = lv_fitnessall + ls_ethnicgroups-fitness.
ENDLOOP.
" 交叉
DO m * 2 TIMES.
LOOP AT ethnicgroups INTO ls_ethnicgroups.
lv_rate = ls_ethnicgroups-fitness / lv_fitnessall * cross.
IF lv_rate = 0.
lv_rate = '0.01'.
ENDIF.
IF calculat_rate( ) <= lv_rate.
IF lt_units_fa IS INITIAL.
lt_units_fa = ls_ethnicgroups-units.
ELSE.
lt_units_mo = ls_ethnicgroups-units.
ENDIF.
ENDIF.
IF lt_units_fa IS NOT INITIAL AND lt_units_mo IS NOT INITIAL AND lt_units_mo <> lt_units_fa.
EXIT.
ENDIF.
ENDLOOP.
IF lt_units_fa IS NOT INITIAL AND lt_units_mo IS NOT INITIAL.
lt_units_ch = unit_cross( units_fa = lt_units_fa units_mo = lt_units_mo ).
IF lt_units_ch IS NOT INITIAL.
CLEAR ls_ethnicgroups.
ls_ethnicgroups-units = lt_units_ch.
ls_ethnicgroups-index = index.
index = index + 1.
APPEND ls_ethnicgroups TO lt_enthicgroups.
ENDIF.
CLEAR:lt_units_fa,lt_units_mo,lt_units_ch.
ENDIF.
ENDDO.
" 变异
LOOP AT ethnicgroups INTO ls_ethnicgroups.
IF calculat_rate( ) <= variation.
unit_var( CHANGING units = ls_ethnicgroups-units ).
ls_ethnicgroups-fitness = calcu_fitness( ls_ethnicgroups-units ).
ENDIF.
MODIFY ethnicgroups FROM ls_ethnicgroups.
ENDLOOP.
LOOP AT lt_enthicgroups INTO ls_ethnicgroups.
IF calculat_rate( ) <= variation.
unit_var( CHANGING units = ls_ethnicgroups-units ).
ENDIF.
ls_ethnicgroups-fitness = calcu_fitness( ls_ethnicgroups-units ).
MODIFY lt_enthicgroups FROM ls_ethnicgroups.
ENDLOOP.
" 记录
CLEAR lt_allgroups.
MOVE-CORRESPONDING ethnicgroups TO lt_allgroups.
ls_allgroups-t = curr_t.
MODIFY lt_allgroups FROM ls_allgroups TRANSPORTING t WHERE units IS NOT INITIAL.
APPEND LINES OF lt_allgroups TO allgroups.
curr_t = curr_t + 1.
CLEAR lt_allgroups.
MOVE-CORRESPONDING lt_enthicgroups TO lt_allgroups.
ls_allgroups-t = curr_t.
MODIFY lt_allgroups FROM ls_allgroups TRANSPORTING t WHERE units IS NOT INITIAL.
APPEND LINES OF lt_allgroups TO allgroups.
APPEND LINES OF lt_enthicgroups TO ethnicgroups.
ENDMETHOD.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Private Method ZCL_GENETIC->RANDOW
* +-------------------------------------------------------------------------------------------------+
* | [--->] LOW TYPE I
* | [--->] HIGH TYPE I
* | [<-()] VALUE TYPE I
* +--------------------------------------------------------------------------------------</SIGNATURE>
METHOD RANDOW.
DATA: lo_random TYPE REF TO cl_abap_random,
lv_seed TYPE i,
lv_range_num TYPE i.
lv_seed = cl_abap_random=>seed( ).
lo_random = cl_abap_random=>create( seed = lv_seed ).
lv_range_num = lo_random->intinrange( low = low high = high ).
value = lv_range_num.
ENDMETHOD.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Private Method ZCL_GENETIC->UNIT_CROSS
* +-------------------------------------------------------------------------------------------------+
* | [--->] UNITS_FA TYPE CHIP_PROPERTY_TAB
* | [--->] UNITS_MO TYPE CHIP_PROPERTY_TAB
* | [<-()] UNITS TYPE CHIP_PROPERTY_TAB
* +--------------------------------------------------------------------------------------</SIGNATURE>
METHOD unit_cross.
DATA: ls_unit LIKE LINE OF units,
ls_unit_fa LIKE LINE OF units,
ls_unit_mo LIKE LINE OF units,
ls_var LIKE LINE OF vars,
lv_low TYPE i,
lv_high TYPE i.
" 数值运算由于限制无法使用二进制交叉,使用中间数模拟交叉结果
DO random_up * 2 TIMES.
LOOP AT units_fa INTO ls_unit_fa.
READ TABLE units_mo INTO ls_unit_mo WITH KEY name = ls_unit_fa-name.
READ TABLE vars INTO ls_var WITH KEY name = ls_unit_fa-name.
lv_low = ls_unit_fa-value.
IF lv_low > ls_unit_mo-value.
lv_low = ls_unit_mo-value.
lv_high = ls_unit_fa-value.
ELSE.
lv_high = ls_unit_mo-value.
ENDIF.
IF calculat_rate( ) < '0.5'.
lv_low = lv_low * 2 - lv_high.
IF lv_low < 0 .
lv_low = 0.
ENDIF.
ELSE.
lv_high = lv_high * 2 - lv_low.
ENDIF.
IF lv_low < ls_var-low.
lv_low = ls_var-low.
ENDIF.
IF lv_high > ls_var-high.
lv_high = ls_var-high.
ENDIF.
ls_unit-value = randow( low = lv_low high = lv_high ).
ls_unit-name = ls_unit_fa-name.
APPEND ls_unit TO units.
CLEAR: ls_unit_mo, ls_unit_fa.
ENDLOOP.
IF check_gen_valid( units ).
EXIT.
ENDIF.
CLEAR units.
ENDDO.
ENDMETHOD.
* <SIGNATURE>---------------------------------------------------------------------------------------+
* | Instance Private Method ZCL_GENETIC->UNIT_VAR
* +-------------------------------------------------------------------------------------------------+
* | [<-->] UNITS TYPE CHIP_PROPERTY_TAB
* +--------------------------------------------------------------------------------------</SIGNATURE>
METHOD unit_var.
DATA: ls_unit LIKE LINE OF units,
lt_units LIKE units,
ls_var LIKE LINE OF vars,
lv_ind TYPE sy-index,
lv_count TYPE i.
lv_count = lines( units ).
lv_ind = randow( low = 1 high = lv_count ).
lt_units = units.
READ TABLE units INTO ls_unit INDEX lv_ind.
IF sy-subrc = 0.
READ TABLE vars INTO ls_var WITH KEY name = ls_unit-name.
IF sy-subrc = 0.
DO random_up TIMES.
ls_unit-value = randow( low = ls_var-low high = ls_var-high ).
IF check_gen_valid( units ).
EXIT.
ENDIF.
CLEAR units.
ENDDO.
ENDIF.
ENDIF.
IF units IS INITIAL.
units = lt_units.
ENDIF.
ENDMETHOD.
ENDCLASS.对象自定义逻辑版本-接口
 对象自定义逻辑版本-示例1源代码
对象自定义逻辑版本-示例1源代码
*----------------------------------------------------------------------*
* 已知
* 0 <= X1 <= 80
* 0 <= X2 <= 40
* 0 <= X3 <= 25
* 15*X1+40*X2+80*X3<=3000
* 求最大值
* X1*20+30*X2+50*X3
* 备注
* X1,X2,X3均为整数,如小数可将限制扩大对应倍数
*----------------------------------------------------------------------*
CLASS lcl_genetic DEFINITION FINAL .
PUBLIC SECTION.
INTERFACES zif_gentic_cus .
PROTECTED SECTION.
PRIVATE SECTION.
ENDCLASS.
CLASS lcl_genetic IMPLEMENTATION.
METHOD zif_gentic_cus~check_gen_valid.
DATA: lv_sum TYPE i.
LOOP AT units INTO DATA(ls_unit).
CASE ls_unit-name.
WHEN 'X1'.
lv_sum = lv_sum + 15 * ls_unit-value.
WHEN 'X2'.
lv_sum = lv_sum + 40 * ls_unit-value.
WHEN 'X3'.
lv_sum = lv_sum + 80 * ls_unit-value.
WHEN OTHERS.
ENDCASE.
ENDLOOP.
IF lv_sum > 3000.
cv_bool = ''.
ELSE.
cv_bool = 'X'.
ENDIF.
ENDMETHOD.
METHOD zif_gentic_cus~calcu_fitness.
DATA: lv_sum TYPE i.
LOOP AT units INTO DATA(ls_unit).
CASE ls_unit-name.
WHEN 'X1'.
lv_sum = lv_sum + 20 * ls_unit-value.
WHEN 'X2'.
lv_sum = lv_sum + 30 * ls_unit-value.
WHEN 'X3'.
lv_sum = lv_sum + 50 * ls_unit-value.
WHEN OTHERS.
ENDCASE.
ENDLOOP.
fitness = lv_sum / 10000.
ENDMETHOD.
ENDCLASS.
DATA: lo_g TYPE REF TO zcl_genetic,
lo_gc TYPE REF TO lcl_genetic,
lt_recs TYPE zcl_genetic_base=>tt_allgroups.
START-OF-SELECTION.
CREATE OBJECT lo_g TYPE zcl_genetic.
CREATE OBJECT lo_gc.
lo_g->init( EXPORTING iv_variation = '0.05'
iv_cross = '0.8'
iv_m = '50'
iv_t = '100'
iv_random_up = '100'
io_genetic_cus = lo_gc
).
lo_g->add_var( name = 'X1' low = 0 high = 80 ).
lo_g->add_var( name = 'X2' low = 0 high = 40 ).
lo_g->add_var( name = 'X3' low = 0 high = 25 ).
DATA(lt_unit) = lo_g->get_result( ).
lo_g->get_record( IMPORTING et_rec = lt_recs ).
WRITE:/ '最优解'.
LOOP AT lt_unit INTO DATA(ls_unit).
WRITE:/ ls_unit-name, ':' ,ls_unit-value.
ENDLOOP.
WRITE:/ '迭代过程'.
SORT lt_recs BY t index.
LOOP AT lt_recs INTO DATA(ls_rec).
WRITE:/ '代数:',ls_rec-t,' 适应度:' ,ls_rec-fitness, ' 值:'.
LOOP AT ls_rec-units INTO ls_unit.
WRITE:ls_unit-name, '-' ,ls_unit-value , ';'.
ENDLOOP.
ENDLOOP.简单装箱问题实践
*----------------------------------------------------------------------*
* 简化装箱单面填充
* 箱 单面80 * 50
* 物料A 30 10 10 共100个
* 物料B 20 10 5 共200个
* 6种摆放方式
* 1 长宽高
* 2 长高宽
* 3 高长宽
* 4 高宽长
* 5 宽高长
* 6 宽长高
* 预置 3个箱子
*----------------------------------------------------------------------*
DATA: boxx TYPE i VALUE '80',
boxy TYPE i VALUE '60',
ax TYPE i VALUE '30',
ay TYPE i VALUE '10',
az TYPE i VALUE '100',
bx TYPE i VALUE '20',
by TYPE i VALUE '10',
bz TYPE i VALUE '5',
amax TYPE i VALUE '100',
bmax TYPE i VALUE '200'.
CLASS lcl_genetic DEFINITION FINAL .
PUBLIC SECTION.
INTERFACES zif_gentic_cus .
PROTECTED SECTION.
PRIVATE SECTION.
ENDCLASS.
CLASS lcl_genetic IMPLEMENTATION.
METHOD zif_gentic_cus~check_gen_valid.
DATA: lv_sumx TYPE i,
lv_sumy TYPE i,
lv_sumz TYPE i.
LOOP AT units INTO DATA(ls_unit_gr) GROUP BY COND string( WHEN strlen( ls_unit_gr-name ) > 3 THEN ls_unit_gr-name+2(1) ELSE '' ).
LOOP AT GROUP ls_unit_gr INTO DATA(ls_unit).
CASE ls_unit-name+0(2).
WHEN 'A1'.
lv_sumx = lv_sumx + ax * ls_unit-value.
lv_sumy = lv_sumy + ay * ls_unit-value.
lv_sumz = lv_sumz + ay * ls_unit-value.
WHEN 'A2'.
lv_sumx = lv_sumx + ax * ls_unit-value.
lv_sumz = lv_sumz + az * ls_unit-value.
lv_sumy = lv_sumy + ay * ls_unit-value.
WHEN 'A3'.
lv_sumz = lv_sumz + az * ls_unit-value.
lv_sumx = lv_sumx + ax * ls_unit-value.
lv_sumy = lv_sumy + ay * ls_unit-value.
WHEN 'A4'.
lv_sumz = lv_sumz + az * ls_unit-value.
lv_sumy = lv_sumy + ay * ls_unit-value.
lv_sumx = lv_sumx + ax * ls_unit-value.
WHEN 'A5'.
lv_sumy = lv_sumy + ay * ls_unit-value.
lv_sumz = lv_sumz + az * ls_unit-value.
lv_sumx = lv_sumx + ax * ls_unit-value.
WHEN 'A6'.
lv_sumy = lv_sumy + ay * ls_unit-value.
lv_sumx = lv_sumx + ax * ls_unit-value.
lv_sumz = lv_sumz + az * ls_unit-value.
WHEN 'B1'.
lv_sumx = lv_sumx + bx * ls_unit-value.
lv_sumy = lv_sumy + by * ls_unit-value.
lv_sumz = lv_sumz + by * ls_unit-value.
WHEN 'B2'.
lv_sumx = lv_sumx + bx * ls_unit-value.
lv_sumz = lv_sumz + bz * ls_unit-value.
lv_sumy = lv_sumy + by * ls_unit-value.
WHEN 'B3'.
lv_sumz = lv_sumz + bz * ls_unit-value.
lv_sumx = lv_sumx + bx * ls_unit-value.
lv_sumy = lv_sumy + by * ls_unit-value.
WHEN 'B4'.
lv_sumz = lv_sumz + bz * ls_unit-value.
lv_sumy = lv_sumy + by * ls_unit-value.
lv_sumx = lv_sumx + bx * ls_unit-value.
WHEN 'B5'.
lv_sumy = lv_sumy + by * ls_unit-value.
lv_sumz = lv_sumz + bz * ls_unit-value.
lv_sumx = lv_sumx + bx * ls_unit-value.
WHEN 'B6'.
lv_sumy = lv_sumy + by * ls_unit-value.
lv_sumx = lv_sumx + bx * ls_unit-value.
lv_sumz = lv_sumz + bz * ls_unit-value.
WHEN OTHERS.
ENDCASE.
ENDLOOP.
IF lv_sumx > 80 OR lv_sumy > 60 OR lv_sumy > 50.
cv_bool = ''.
ELSE.
cv_bool = 'X'.
ENDIF.
ENDLOOP.
ENDMETHOD.
METHOD zif_gentic_cus~calcu_fitness.
DATA: lv_sum TYPE i,
lv_suma TYPE i,
lv_r TYPE i.
LOOP AT units INTO DATA(ls_unit_gr) GROUP BY ( name+2(1) = ls_unit_gr-name+2(1) ).
lv_r = ls_unit_gr-name+2(1).
CLEAR lv_suma.
LOOP AT GROUP ls_unit_gr INTO DATA(ls_unit).
lv_suma = lv_suma + ls_unit-value.
ENDLOOP.
lv_sum = lv_sum + lv_suma * lv_r.
ENDLOOP.
fitness = lv_sum / 10000.
ENDMETHOD.
ENDCLASS.
DATA: lo_g TYPE REF TO zcl_genetic,
lo_gc TYPE REF TO lcl_genetic,
lt_recs TYPE zcl_genetic_base=>tt_allgroups.
START-OF-SELECTION.
CREATE OBJECT lo_g TYPE zcl_genetic.
CREATE OBJECT lo_gc.
lo_g->init( EXPORTING iv_variation = '0.05'
iv_cross = '0.8'
iv_m = '50'
iv_t = '100'
iv_random_up = '100'
io_genetic_cus = lo_gc
).
lo_g->add_var( name = 'A11' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV ax ) * ( boxy DIV ay ) ) ).
lo_g->add_var( name = 'A21' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV ax ) * ( boxy DIV az ) ) ).
lo_g->add_var( name = 'A31' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV az ) * ( boxy DIV ax ) ) ).
lo_g->add_var( name = 'A41' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV az ) * ( boxy DIV ay ) ) ).
lo_g->add_var( name = 'A51' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV ay ) * ( boxy DIV az ) ) ).
lo_g->add_var( name = 'A61' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV ay ) * ( boxy DIV ax ) ) ).
lo_g->add_var( name = 'B11' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV bx ) * ( boxy DIV by ) ) ).
lo_g->add_var( name = 'B21' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV bx ) * ( boxy DIV bz ) ) ).
lo_g->add_var( name = 'B31' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV bz ) * ( boxy DIV bx ) ) ).
lo_g->add_var( name = 'B41' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV bz ) * ( boxy DIV by ) ) ).
lo_g->add_var( name = 'B51' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV by ) * ( boxy DIV bz ) ) ).
lo_g->add_var( name = 'B61' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV by ) * ( boxy DIV bx ) ) ).
lo_g->add_var( name = 'A12' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV ax ) * ( boxy DIV ay ) ) ).
lo_g->add_var( name = 'A22' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV ax ) * ( boxy DIV az ) ) ).
lo_g->add_var( name = 'A32' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV az ) * ( boxy DIV ax ) ) ).
lo_g->add_var( name = 'A42' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV az ) * ( boxy DIV ay ) ) ).
lo_g->add_var( name = 'A52' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV ay ) * ( boxy DIV az ) ) ).
lo_g->add_var( name = 'A62' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV ay ) * ( boxy DIV ax ) ) ).
lo_g->add_var( name = 'B12' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV bx ) * ( boxy DIV by ) ) ).
lo_g->add_var( name = 'B22' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV bx ) * ( boxy DIV bz ) ) ).
lo_g->add_var( name = 'B32' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV bz ) * ( boxy DIV bx ) ) ).
lo_g->add_var( name = 'B42' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV bz ) * ( boxy DIV by ) ) ).
lo_g->add_var( name = 'B52' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV by ) * ( boxy DIV bz ) ) ).
lo_g->add_var( name = 'B62' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV by ) * ( boxy DIV bx ) ) ).
lo_g->add_var( name = 'A13' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV ax ) * ( boxy DIV ay ) ) ).
lo_g->add_var( name = 'A23' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV ax ) * ( boxy DIV az ) ) ).
lo_g->add_var( name = 'A33' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV az ) * ( boxy DIV ax ) ) ).
lo_g->add_var( name = 'A43' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV az ) * ( boxy DIV ay ) ) ).
lo_g->add_var( name = 'A53' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV ay ) * ( boxy DIV az ) ) ).
lo_g->add_var( name = 'A63' low = 0 high = nmax( val1 = amax val2 = ( boxx DIV ay ) * ( boxy DIV ax ) ) ).
lo_g->add_var( name = 'B13' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV bx ) * ( boxy DIV by ) ) ).
lo_g->add_var( name = 'B23' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV bx ) * ( boxy DIV bz ) ) ).
lo_g->add_var( name = 'B33' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV bz ) * ( boxy DIV bx ) ) ).
lo_g->add_var( name = 'B43' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV bz ) * ( boxy DIV by ) ) ).
lo_g->add_var( name = 'B53' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV by ) * ( boxy DIV bz ) ) ).
lo_g->add_var( name = 'B63' low = 0 high = nmax( val1 = bmax val2 = ( boxx DIV by ) * ( boxy DIV bx ) ) ).
DATA(lt_unit) = lo_g->get_result( ).
lo_g->get_record( IMPORTING et_rec = lt_recs ).
WRITE:/ '最优解'.
LOOP AT lt_unit INTO DATA(ls_unit).
WRITE:/ ls_unit-name, ':' ,ls_unit-value.
ENDLOOP.
WRITE:/ '迭代过程'.
SORT lt_recs BY t index.
LOOP AT lt_recs INTO DATA(ls_rec).
WRITE:/ '代数:',ls_rec-t,' 适应度:' ,ls_rec-fitness, ' 值:'.
LOOP AT ls_rec-units INTO ls_unit.
WRITE:ls_unit-name, '-' ,ls_unit-value , ';'.
ENDLOOP.
ENDLOOP.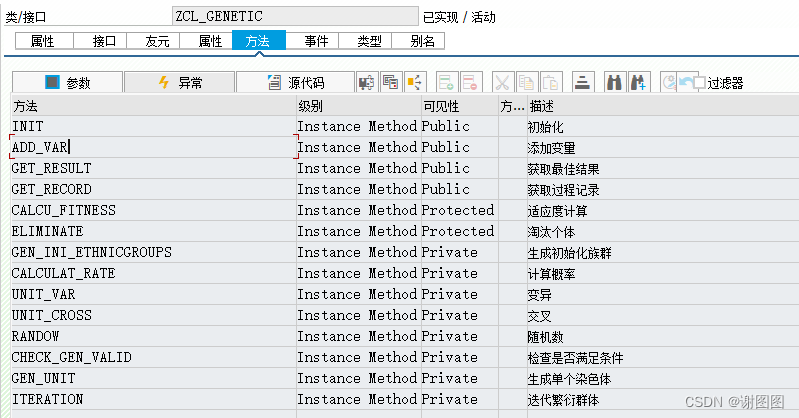
![Navicat连接SQLSever报错:[08001] MicrosoftTCP Provider 远程主机强迫关闭了一个现有的连接](https://img-blog.csdnimg.cn/direct/3042b73736dc460eb532fe730002166b.png)