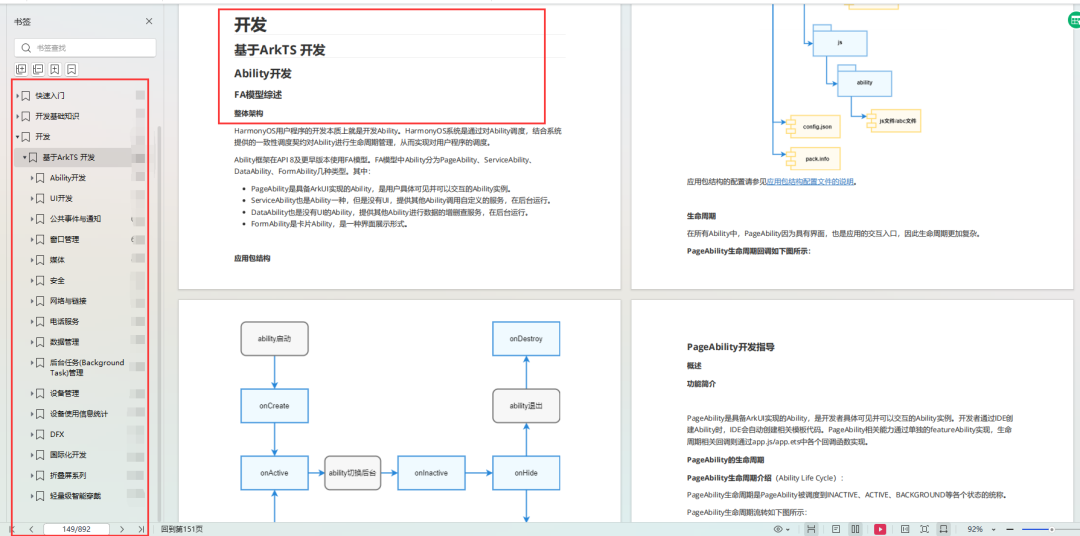
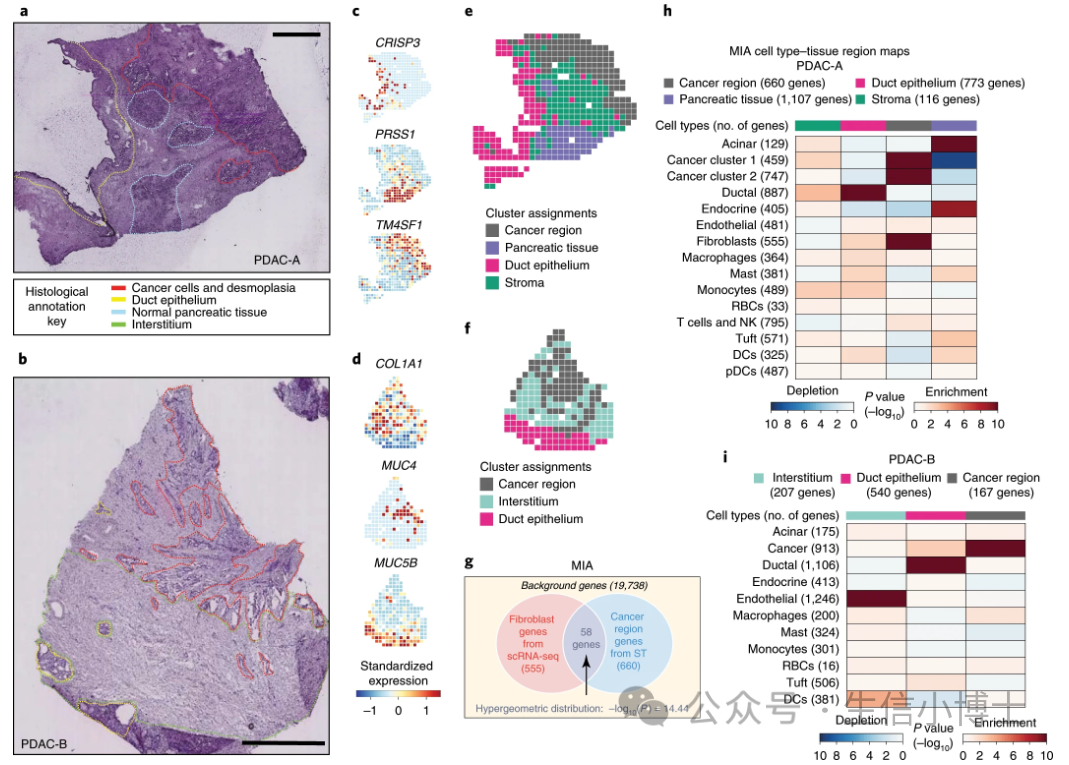
原文:Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas | Nature Biotechnology
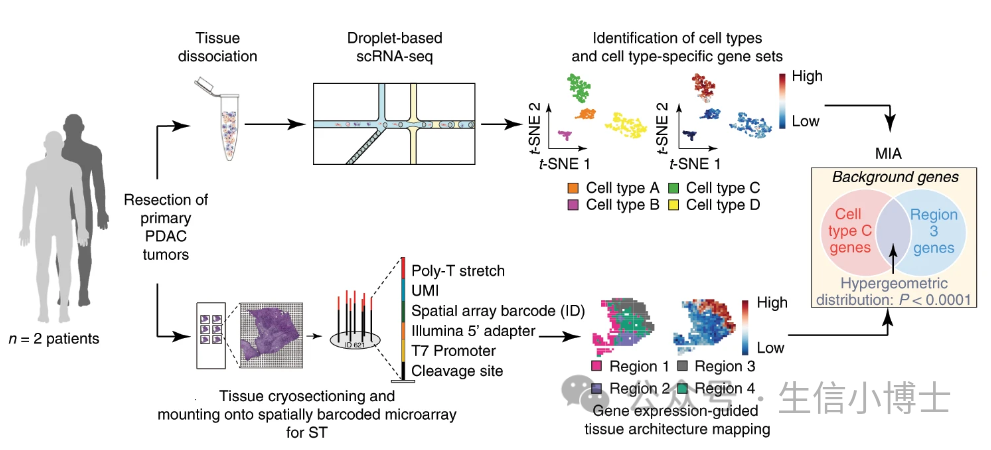
研究者采用 MIA 联合 scRNAseq 和 ST 数据,分析原发性胰腺导管腺癌(PDAC)的组织结构。大家好,
MIA 首先使用以下步骤来描述细胞类型特异性基因集和组织区域特异性基因集:
-
在 scRNAseq 数据中,识别每个细胞类型中比其余的细胞都高表达的基因,定义细胞类型基因集。
-
基于 ST 数据,确定各个空间区域相对于其他区域有显著高表达的基因集(空间区域基因集)。
-
然后,MIA 计算每一对细胞类型基因集的特异性和区域特异性,并采用超几何分布来评估显著的富集或耗竭。
可以看到MIA需要我们提供 单细胞marker基因集 和 空间marker基因基因集合(最好是同一套样本同时做了空转和单细胞,如果不是同一套,可能对生物学解释有些影响,但是不影响生信分析流程)
完整代码,放在文末,这里分别说下所需要的单细胞和空转输入数据
空转数据输入格式
> ####st-----> cluster_gene_stat=FindAllMarkers(d.all,only.pos = TRUE,logfc.threshold = 0.9)Calculating cluster cluster2|++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=00sCalculating cluster cluster4|++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=01sCalculating cluster cluster1|++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=00sCalculating cluster cluster3|++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=03sCalculating cluster cluster5|++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=00sCalculating cluster cluster0|++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=00sCalculating cluster cluster7Calculating cluster cluster6|++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=02sCalculating cluster cluster8|++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed=00s> head(cluster_gene_stat)p_val avg_log2FC pct.1 pct.2 p_val_adj cluster geneMarco 1.168778e-259 1.9388057 0.874 0.298 2.268598e-255 cluster2 MarcoWfdc21 1.246929e-191 1.5790076 0.836 0.361 2.420289e-187 cluster2 Wfdc21Chil3 2.075188e-149 1.7852619 0.995 0.946 4.027939e-145 cluster2 Chil3Plet1 2.197103e-129 1.2324047 0.870 0.518 4.264576e-125 cluster2 Plet1Ccl6 8.553776e-111 1.2698564 1.000 0.949 1.660288e-106 cluster2 Ccl6Ly6i 9.178082e-100 0.9324478 0.972 0.550 1.781466e-95 cluster2 Ly6i> table(cluster_gene_stat$cluster)cluster2 cluster4 cluster1 cluster3 cluster5 cluster0 cluster7 cluster6 cluster87 142 31 247 7 10 0 120 7> # Initialize a dataframe for us to store values in:> st.clusts=paste0("cluster",seq(0,8,1))> head(cluster_gene_stat)p_val avg_log2FC pct.1 pct.2 p_val_adj cluster geneMarco 1.168778e-259 1.9388057 0.874 0.298 2.268598e-255 cluster2 MarcoWfdc21 1.246929e-191 1.5790076 0.836 0.361 2.420289e-187 cluster2 Wfdc21Chil3 2.075188e-149 1.7852619 0.995 0.946 4.027939e-145 cluster2 Chil3Plet1 2.197103e-129 1.2324047 0.870 0.518 4.264576e-125 cluster2 Plet1Ccl6 8.553776e-111 1.2698564 1.000 0.949 1.660288e-106 cluster2 Ccl6Ly6i 9.178082e-100 0.9324478 0.972 0.550 1.781466e-95 cluster2 Ly6i> st.marker.list=split(cluster_gene_stat,f = cluster_gene_stat$cluster)> st.marker.list = lapply(st.marker.list,rownames)> head(st.marker.list)$cluster2[1] "Marco" "Wfdc21" "Chil3" "Plet1" "Ccl6" "Ly6i" "Ctsk"$cluster4[1] "Cdhr4" "Rsph1" "Gabrp" "Ccdc153" "Dnah5" "Foxj1" "1110017D15Rik" "Lrrc23"[9] "Cfap43" "Cdhr3" "Dnali1" "Pifo" "Scgb1a1" "Ak7" "Tmem212" "Gm19935"[17] "Cyp2f2" "Dnah6" "Cfap65" "1700007K13Rik" "Fam183b" "Cyp2s1" "Cfap206" "Nme9"[25] "1700016K19Rik" "Mgst1" "BC051019" "AU040972" "Wfdc2" "Meig1" "Fmo3" "Cfap126"[33] "Mapk15" "Tctex1d4" "Acta2" "Cckar" "Drc3" "Scgb3a2" "Rbp4" "Dynlrb2"[41] "Cldn10" "B430010I23Rik" "Riiad1" "3300002A11Rik" "Nme5" "Ldhb" "1700024G13Rik" "Hp"[49] "Cbr2" "Tspan1" "Anxa8" "Odf3b" "Cnn1" "Ccdc17" "Mlf1" "Dcn"[57] "Mgat3" "Cxcl17" "Slc16a11" "Gpx2" "Chad" "Clic6" "5330417C22Rik" "Gsta4"[65] "Retnla" "Myh11" "Calml4" "Cd55" "Cygb" "1700094D03Rik" "Cd24a" "Alas1"[73] "Actg2" "Mmp2" "Lypd2" "Aldh1a7" "Myl9" "Tpm2" "1700088E04Rik" "Tubb4b"[81] "Lrrc51" "Elof1" "C4b" "Vpreb3" "Igfbp5" "Ccl21a" "Arhgdig" "Pon1"[89] "Igfbp4" "Csrp2" "Map1b" "Chchd10" "Endog" "Col14a1" "Tagln" "Por"[97] "Nupr1" "Dcxr" "Cyp2a5" "Pcp4l1" "Mustn1" "Net1" "Traf3ip1" "Fbln1"[105] "Aebp1" "Selenbp1" "Enah" "Adamts2" "Des" "Pi16" "Gsto1" "Ltf"[113] "Muc5b" "Actc1" "Aox3" "Ces1d" "Tmem107" "Bphl" "Serpinf1" "C3"[121] "Tst" "Tff2" "Pigr" "Aldh1a1" "Scgb1c1" "Cpe" "Aldh3b1" "Tppp3"[129] "Gstm1" "Clec3b" "Scgb3a1" "Gsta3" "Bpifa1" "Bpifb1" "Igha" "Reg3g"[137] "Igfbp6" "Gsn" "Jchain" "Igkc" "Iglc1" "Cfd"$cluster1[1] "Lcn2" "Saa3" "Lrg1" "Ccl2" "Sftpd" "Chil1" "Slc26a4" "Ctsc" "Itih4" "Slpi" "Il33" "Fabp5" "Hc"[14] "Ly6i1" "Cd74" "Clu" "Fasn" "Muc1" "Spp1" "Igha1" "Igkc1" "Ccl7" "Ctss" "Wfdc17" "Jchain1" "Ptgs1"[27] "Npc2" "Tgfbi" "Ighg2c" "Ighg2b" "Ighg1"$cluster3[1] "Mmp12" "Clec4n" "Gpnmb" "Lgals3" "Lilr4b" "Prdx1" "Lilrb4a" "Ccl9" "Psap" "Clec4d" "Clec4e"[12] "Itgb2" "Il1rn" "Ftl1" "Lhfpl2" "Ltbp2" "Fth1" "Ctsb" "Cstb" "S100a4" "Pla2g7" "Ctss1"[23] "Spp11" "Ctsd" "Capg" "Atp6v0c" "Cd68" "Ctsz" "Cyba" "Tyrobp" "Cd63" "Litaf" "Tcirg1"[34] "Bcl2a1b" "Cybb" "Esd" "AA467197" "Slc7a11" "Fn1" "Hvcn1" "Fcer1g" "Lpl" "Cd52" "Ftl1-ps1"[45] "Lipa" "Csf2rb" "Fabp51" "Fcgr2b" "Saa31" "Pld3" "Fam20c" "Mpeg1" "Trem2" "Col3a1" "Ccl61"[56] "Pgam1" "Tmsb10" "Acod1" "Adam8" "Sirpa" "Plin2" "Pkm" "Col5a2" "Itgax" "F10" "Creg1"[67] "Slc11a1" "Arpc1b" "Sh3bgrl3" "Igf1" "Grn" "Eif4a1" "Slc15a3" "Ccl3" "Col1a1" "Rnf128" "Lyz2"[78] "Col1a2" "Serpine2" "Txn1" "Wfdc171" "H2-D1" "Mmp19" "Lgmn" "Eln" "Sfrp1" "Ctsa" "Rnh1"[89] "Tnfrsf1b" "Tnfaip2" "Fxyd5" "Rgs1" "Pgd" "Cd53" "Timp1" "Ifi30" "F7" "Ccl21" "Ncf2"[100] "Ninj1" "Fcgr3" "Lgals1" "Vmp1" "Lcp1" "Msr1" "Anxa1" "Gapdh" "Gpx1" "Csf2ra" "Col5a1"[111] "Atox1" "Anpep" "Prkcd" "Atp6v1e1" "Col4a1" "Sulf1" "Ctsl" "Hexa" "Atp6v0d2" "Cd44" "Rilpl2"[122] "Gusb" "Sgk1" "Plaur" "Cd741" "Igfbp7" "Bhlhe40" "Csf2rb2" "Mdk" "Atp6v0b" "Cotl1" "Ptafr"[133] "Msrb1" "Mgp" "Fbn1" "Spi1" "Ppic" "Lat2" "Plek" "B4galt5" "Gpr137b" "Laptm5" "Mfap5"[144] "Itgam" "Cxcl2" "Ncf1" "Cd9" "Tlr2" "Txnrd1" "Acp5" "Tnc" "Mmp21" "Fndc1" "Junb"[155] "Psmd8" "Marcksl1" "Atp6v1b2" "Cd300c2" "Tuba1c" "Emp3" "Cd84" "Fstl1" "Pirb" "Slfn2" "Card19"[166] "Slpi1" "Basp1" "Aprt" "Bst2" "Hmox1" "Prelid1" "Bst1" "Mfsd12" "Cxcl12" "Myo1f" "Sgpl1"[177] "Rhoc" "Col4a2" "Apoe" "Glipr1" "Sh3pxd2b" "Sbno2" "Mif" "Myo5a" "Plxnd1" "Clic1" "Socs3"[188] "Alox5ap" "Ctsk1" "Lgals3bp" "Ncf4" "Blvrb" "Thbs2" "Cyth4" "Fmod" "Cpxm1" "Srxn1" "Ms4a6d"[199] "Pycard" "Tgfbi1" "Prdx5" "Cd14" "Cxcl10" "Ctsh" "Col18a1" "Ccl71" "Clu1" "Cndp2" "Atp6v1c1"[210] "Eno1" "Pmepa1" "Colgalt1" "Cilp" "Bmp1" "Unc93b1" "Col6a3" "Mt1" "Sema4d" "Isg15" "C1qtnf6"[221] "Aebp11" "Ccdc80" "Il1b" "Npc21" "Grem1" "Iigp1" "Ccl4" "Nrep" "S100a8" "C1qb" "Man2b1"[232] "Abca1" "Igha2" "Irf7" "Cxcl9" "Igkc2" "Jchain2" "Slfn4" "C1qa" "C1qc" "Ifit3" "Rsad2"[243] "S100a9" "Ighg2b1" "Ifit1" "Ighg2c1" "Ighg11"$cluster5[1] "Scgb3a21" "Cyp2f21" "Inmt" "Hbb-bs" "Hba-a2" "Hba-a1" "Hbb-bt"$cluster0[1] "Inmt1" "Hba-a11" "Hbb-bs1" "Hbb-bt1" "Hba-a21" "Fmo2" "Pcolce2" "Cyp4b1" "Tspan7" "Hpgd"
###########################
最终得到空转中的数据,整理出下面两个对象
st.clusts,是一个向量
st.marker.list,是一个列表,元素就是每个区域的marker基因,列表的names就构成了st.lusts
st.clusts[1] "cluster0" "cluster1" "cluster2" "cluster3" "cluster4" "cluster5" "cluster6" "cluster7" "cluster8"> st.marker.list$cluster2[1] "Marco" "Wfdc21" "Chil3" "Plet1" "Ccl6" "Ly6i" "Ctsk"$cluster4[1] "Cdhr4" "Rsph1" "Gabrp" "Ccdc153" "Dnah5" "Foxj1" "1110017D15Rik" "Lrrc23"[9] "Cfap43" "Cdhr3" "Dnali1" "Pifo" "Scgb1a1" "Ak7" "Tmem212" "Gm19935"[17] "Cyp2f2" "Dnah6" "Cfap65" "1700007K13Rik" "Fam183b" "Cyp2s1" "Cfap206" "Nme9"[25] "1700016K19Rik" "Mgst1" "BC051019" "AU040972" "Wfdc2" "Meig1" "Fmo3" "Cfap126"[33] "Mapk15" "Tctex1d4" "Acta2" "Cckar" "Drc3" "Scgb3a2" "Rbp4" "Dynlrb2"[41] "Cldn10" "B430010I23Rik" "Riiad1" "3300002A11Rik" "Nme5" "Ldhb" "1700024G13Rik" "Hp"[49] "Cbr2" "Tspan1" "Anxa8" "Odf3b" "Cnn1" "Ccdc17" "Mlf1" "Dcn"[57] "Mgat3" "Cxcl17" "Slc16a11" "Gpx2" "Chad" "Clic6" "5330417C22Rik" "Gsta4"[65] "Retnla" "Myh11" "Calml4" "Cd55" "Cygb" "1700094D03Rik" "Cd24a" "Alas1"[73] "Actg2" "Mmp2" "Lypd2" "Aldh1a7" "Myl9" "Tpm2" "1700088E04Rik" "Tubb4b"[81] "Lrrc51" "Elof1" "C4b" "Vpreb3" "Igfbp5" "Ccl21a" "Arhgdig" "Pon1"[89] "Igfbp4" "Csrp2" "Map1b" "Chchd10" "Endog" "Col14a1" "Tagln" "Por"[97] "Nupr1" "Dcxr" "Cyp2a5" "Pcp4l1" "Mustn1" "Net1" "Traf3ip1" "Fbln1"[105] "Aebp1" "Selenbp1" "Enah" "Adamts2" "Des" "Pi16" "Gsto1" "Ltf"[113] "Muc5b" "Actc1" "Aox3" "Ces1d" "Tmem107" "Bphl" "Serpinf1" "C3"[121] "Tst" "Tff2" "Pigr" "Aldh1a1" "Scgb1c1" "Cpe" "Aldh3b1" "Tppp3"[129] "Gstm1" "Clec3b" "Scgb3a1" "Gsta3" "Bpifa1" "Bpifb1" "Igha" "Reg3g"[137] "Igfbp6" "Gsn" "Jchain" "Igkc" "Iglc1" "Cfd"$cluster1[1] "Lcn2" "Saa3" "Lrg1" "Ccl2" "Sftpd" "Chil1" "Slc26a4" "Ctsc" "Itih4" "Slpi" "Il33" "Fabp5" "Hc"[14] "Ly6i1" "Cd74" "Clu" "Fasn" "Muc1" "Spp1" "Igha1" "Igkc1" "Ccl7" "Ctss" "Wfdc17" "Jchain1" "Ptgs1"[27] "Npc2" "Tgfbi" "Ighg2c" "Ighg2b" "Ighg1"$cluster3[1] "Mmp12" "Clec4n" "Gpnmb" "Lgals3" "Lilr4b" "Prdx1" "Lilrb4a" "Ccl9" "Psap" "Clec4d" "Clec4e"[12] "Itgb2" "Il1rn" "Ftl1" "Lhfpl2" "Ltbp2" "Fth1" "Ctsb" "Cstb" "S100a4" "Pla2g7" "Ctss1"[23] "Spp11" "Ctsd" "Capg" "Atp6v0c" "Cd68" "Ctsz" "Cyba" "Tyrobp" "Cd63" "Litaf" "Tcirg1"[34] "Bcl2a1b" "Cybb" "Esd" "AA467197" "Slc7a11" "Fn1" "Hvcn1" "Fcer1g" "Lpl" "Cd52" "Ftl1-ps1"[45] "Lipa" "Csf2rb" "Fabp51" "Fcgr2b" "Saa31" "Pld3" "Fam20c" "Mpeg1" "Trem2" "Col3a1" "Ccl61"[56] "Pgam1" "Tmsb10" "Acod1" "Adam8" "Sirpa" "Plin2" "Pkm" "Col5a2" "Itgax" "F10" "Creg1"[67] "Slc11a1" "Arpc1b" "Sh3bgrl3" "Igf1" "Grn" "Eif4a1" "Slc15a3" "Ccl3" "Col1a1" "Rnf128" "Lyz2"[78] "Col1a2" "Serpine2" "Txn1" "Wfdc171" "H2-D1" "Mmp19" "Lgmn" "Eln" "Sfrp1" "Ctsa" "Rnh1"[89] "Tnfrsf1b" "Tnfaip2" "Fxyd5" "Rgs1" "Pgd" "Cd53" "Timp1" "Ifi30" "F7" "Ccl21" "Ncf2"[100] "Ninj1" "Fcgr3" "Lgals1" "Vmp1" "Lcp1" "Msr1" "Anxa1" "Gapdh" "Gpx1" "Csf2ra" "Col5a1"[111] "Atox1" "Anpep" "Prkcd" "Atp6v1e1" "Col4a1" "Sulf1" "Ctsl" "Hexa" "Atp6v0d2" "Cd44" "Rilpl2"[122] "Gusb" "Sgk1" "Plaur" "Cd741" "Igfbp7" "Bhlhe40" "Csf2rb2" "Mdk" "Atp6v0b" "Cotl1" "Ptafr"[133] "Msrb1" "Mgp" "Fbn1" "Spi1" "Ppic" "Lat2" "Plek" "B4galt5" "Gpr137b" "Laptm5" "Mfap5"[144] "Itgam" "Cxcl2" "Ncf1" "Cd9" "Tlr2" "Txnrd1" "Acp5" "Tnc" "Mmp21" "Fndc1" "Junb"[155] "Psmd8" "Marcksl1" "Atp6v1b2" "Cd300c2" "Tuba1c" "Emp3" "Cd84" "Fstl1" "Pirb" "Slfn2" "Card19"[166] "Slpi1" "Basp1" "Aprt" "Bst2" "Hmox1" "Prelid1" "Bst1" "Mfsd12" "Cxcl12" "Myo1f" "Sgpl1"[177] "Rhoc" "Col4a2" "Apoe" "Glipr1" "Sh3pxd2b" "Sbno2" "Mif" "Myo5a" "Plxnd1" "Clic1" "Socs3"[188] "Alox5ap" "Ctsk1" "Lgals3bp" "Ncf4" "Blvrb" "Thbs2" "Cyth4" "Fmod" "Cpxm1" "Srxn1" "Ms4a6d"[199] "Pycard" "Tgfbi1" "Prdx5" "Cd14" "Cxcl10" "Ctsh" "Col18a1" "Ccl71" "Clu1" "Cndp2" "Atp6v1c1"[210] "Eno1" "Pmepa1" "Colgalt1" "Cilp" "Bmp1" "Unc93b1" "Col6a3" "Mt1" "Sema4d" "Isg15" "C1qtnf6"[221] "Aebp11" "Ccdc80" "Il1b" "Npc21" "Grem1" "Iigp1" "Ccl4" "Nrep" "S100a8" "C1qb" "Man2b1"[232] "Abca1" "Igha2" "Irf7" "Cxcl9" "Igkc2" "Jchain2" "Slfn4" "C1qa" "C1qc" "Ifit3" "Rsad2"[243] "S100a9" "Ighg2b1" "Ifit1" "Ighg2c1" "Ighg11"$cluster5[1] "Scgb3a21" "Cyp2f21" "Inmt" "Hbb-bs" "Hba-a2" "Hba-a1" "Hbb-bt"$cluster0[1] "Inmt1" "Hba-a11" "Hbb-bs1" "Hbb-bt1" "Hba-a21" "Fmo2" "Pcolce2" "Cyp4b1" "Tspan7" "Hpgd"$cluster7character(0)$cluster6[1] "Kcnj3" "Myl1" "Mybphl" "Tmem182" "Trdn" "Art1" "Myom2" "Hamp" "Tbx20" "Hrc" "Fndc5"[12] "Eef1a2" "Casq2" "Sbk3" "Gm15543" "Ank1" "Pgam2" "Ryr2" "Smpx" "Txlnb" "Srl" "Myoz2"[23] "Obscn" "Ldb3" "Sln" "Ttn" "Smyd1" "Csrp3" "Ckmt2" "Actn2" "Ckm" "Cox8b" "Hspb7"[34] "Myl7" "Tcap" "Myl4" "Tnnc1" "Tnni3" "Cmya5" "Pln" "Mb" "Tnnt2" "Myh6" "Cox7a1"[45] "Cox6a2" "Fabp3" "Actc11" "Atp2a2" "Ankrd1" "Tnnt1" "Myl3" "Slc2a4" "Mybpc3" "Aldh1b1" "Scara5"[56] "Nppa" "Fhl2" "Coq8a" "Popdc2" "Cpe1" "Myom1" "mt-Co3" "Dcn1" "Tpm1" "mt-Co2" "Pi161"[67] "Eno3" "Pam" "Slc25a4" "Pde4dip" "mt-Co1" "mt-Nd1" "Asb2" "mt-Nd3" "Car3" "mt-Nd4" "mt-Cytb"[78] "Got1" "Pygm" "Pfkm" "Ndrg2" "Cfd1" "mt-Nd2" "Igfbp51" "Hspb6" "mt-Atp6" "Cfh" "Fxyd6"[89] "Fbln11" "Tmem38a" "Clec3b1" "Gsn1" "Hadhb" "Art3" "Htra3" "Mdh1" "Atp5b" "Atp5g1" "Lum"[100] "Aco2" "Pdlim5" "Col14a11" "Dsp" "Mdh2" "Acsl1" "Cryab" "mt-Nd5" "Fxyd1" "Idh3a" "Pdha1"[111] "Prss23" "Uqcrfs1" "Cycs" "Got2" "Acadm" "Mmrn1" "F13a1" "Fabp4" "Ccl8" "Ccl21a1"$cluster8[1] "Myl71" "Sln1" "Myl41" "Myh61" "Tnnt21" "Tnni31" "Tcap1"
单细胞数据输入格式
单细胞中,整理出下面两个对象
sc.clusts,单细胞的每个细胞类型名称,是个向量
cellType_marker,单细胞的marker基因,列表形式,其names就是sc.clusts
> sc.clusts[1] "Inmt fibroblast" "Hhip fibroblast" "Universal fibroblast" "Grem1 fibroblast" "Myofibroblast"[6] "Monocyte" "AM3" "AM2" "NK cell" "AM1"[11] "Dendritic cell" "Endothelial cell-2" "Cycling basal cell" "B cell" "Neutrophil"[16] "T cell" "IM" "AT2 cell-2" "Igha+ AT2 cell" "Endothelial cell-1"[21] "AT1 cell" "Clara cell" "Ciliated cell" "AT2 cell-1" "Ig-producing B cell"> cellType_marker$`Inmt fibroblast`[1] "Inmt" "Gpx3" "Sod3" "Fmo2" "Serping1" "Bgn" "Mfap4" "Pcolce2" "Sparcl1" "Npnt"[11] "Ccn1" "Limch1" "Fhl1" "Ogn" "Ltbp4" "Clec3b" "Rbp1" "Timp3" "Col1a2" "Mxra8"[21] "Mettl7a1" "Prelp" "Cxcl14" "Plpp3" "Ptgis" "Plxdc2" "Cfh" "Col1a1" "Col3a1" "Aldh1a1"[31] "Tcf21" "C7" "Ppp1r14a" "Rarres2" "Pdgfra" "Olfml3" "Fxyd1" "Nbl1" "Cdo1" "Adh1"[41] "Mylk" "Pcolce" "Sept4" "Selenbp1" "Serpinh1" "Serpine2" "Spon1" "Col6a1" "Hspb1" "Gpc3"[51] "Dpt" "Nfib" "Cald1" "Colec12" "Loxl1" "Col6a2" "Cp" "Itga8" "C1s1" "Serpina3n"[61] "Atp1a2" "Crispld2" "Dpep1" "Efemp1" "Gpm6b" "Prex2" "Nr2f2" "Ces1d" "Myh10" "Rcn3"[71] "Gas6" "Fbln1" "Mamdc2" "Gsta3" "Mfap5" "Fkbp9" "Mmp2" "Vcam1" "Col6a3" "Ms4a4d"[81] "Abca8a" "Tnxb" "C1ra" "Cryab" "Slc38a5" "Nebl" "Scarf2" "Vegfd" "Nox4" "F2r"[91] "Sdc2" "Col13a1" "Gstt1" "Palld" "Ccn5" "Cpq" "Tmem204" "Cdh11" "Myo1b" "Ppic"[101] "Sparc" "Dbp" "Gstm2" "Rnase4" "Gng11" "Selenom" "C4b" "Mgp" "Igfbp7" "Hsd11b1"[111] "Fermt2" "Csrp1" "Slc43a3" "Nedd4" "Mfge8" "Tns1" "Cd81" "Tmem176a" "Nupr1" "Id3"[121] "Tmem176b" "Pmp22" "Lbh" "Tpm1" "Igfbp6" "Gsn" "Ecm1" "Klf9" "Zbtb20" "Oat"[131] "Gyg" "Dpysl2" "Timp2" "Cebpd" "Macf1" "Apoe" "Nenf" "Sptbn1" "Tuba1a" "C3"[141] "Egr1" "Aldh2" "Gstm1" "Ctsl" "Jun"$`Hhip fibroblast`[1] "Hhip" "Enpp2" "Aspn" "Igfbp3" "Cyp2e1" "Pcsk5" "Cdh4" "Sgcg"[9] "Mustn1" "Grem2" "6330403K07Rik" "Lum" "Sod3" "Rarres2" "Ltbp3" "Igfbp5"[17] "Ltbp4" "Prelp" "Tnxb" "Tagln" "Ddr2" "Des" "Fxyd1" "Gm13889"[25] "Serpine2" "Ogn" "Dpt" "Auts2" "Tpm2" "Lama4" "Bgn" "Ptgis"[33] "Atp1a2" "Fmo2" "Sdc2" "Tgfbr3" "Col1a1" "Col6a3" "Col1a2" "Mdk"[41] "Serping1" "Col3a1" "Palld" "Nfib" "Olfml2b" "Myl9" "Mfap5" "Sparcl1"[49] "Timp3" "Igfbp7" "Crispld2" "Fstl1" "Rcn3" "Pam" "Prnp" "Mylk"[57] "Gpx3" "Acta2" "Serpinh1" "Cavin3" "Mgp" "Sparc" "Cd81" "P2ry14"[65] "Dcn" "Tmem176b" "Hmcn1" "Igf1" "Selenom" "Adamts10" "Cald1" "Zbtb20"[73] "Nedd4" "Csrp1" "Nfia" "Klf9" "Gsn" "Crip2" "Tmem176a" "Nfix"[81] "Tpm1" "Sdc4" "Nenf" "Ctsl" "Nupr1" "Lpp" "Dst" "Gstm1"[89] "Id3" "Laptm4a" "Apoe" "Thbs1" "Cebpd"$`Universal fibroblast`[1] "Dcn" "Clec3b" "Mgp" "Serping1" "Inmt" "Col1a2" "Igfbp7" "Timp3" "Sparc" "Igfbp4"[11] "Col3a1" "Pcolce2" "Col1a1" "Bgn" "Igfbp6" "Aebp1" "Dpt" "Fbln1" "Ltbp4" "Mfap5"[21] "Cfh" "Cygb" "Plpp3" "Rarres2" "Serpinf1" "Gpc3" "Sparcl1" "Rbp1" "Nbl1" "Sod3"[31] "C1s1" "Pcolce" "Ccn1" "Ogn" "Selenom" "Dpep1" "Fmo2" "Serpinh1" "Adh1" "Fxyd1"[41] "C1ra" "Fstl1" "Cd34" "Gpx3" "Rnase4" "Rcn3" "Nfib" "Plxdc2" "Mmp2" "Rbp4"[51] "Abi3bp" "C4b" "Abca8a" "Col6a2" "Ptgis" "Tnxb" "Akap12" "Mfap4" "Fhl1" "Col14a1"[61] "Col6a1" "Gas6" "Slc43a3" "Adamts2" "C7" "Loxl1" "Cavin3" "Gas1" "Ddr2" "Efemp1"[71] "Mxra8" "Bicc1" "Ccl11" "Htra3" "Gstm2" "Fbn1" "Entpd2" "Pdgfra" "Mmp23" "Cxcl12"[81] "Meg3" "Cpq" "Tcf21" "Prelp" "Timp1" "Lhfp" "Rbms3" "Scara5" "Cald1" "Ccdc80"[91] "Csrp2" "Pam" "Crtap" "Pmp22" "Nedd4" "Ecm1" "Id3" "Cd81" "Nenf" "Timp2"[101] "Pi16" "Nfix" "Vkorc1" "Il11ra1" "Nupr1" "C3" "Gsn" "Pdlim2" "Zbtb20" "Cd302"[111] "Cebpd" "Serpinb6a" "Tuba1a" "Klf9" "Ctsl" "Mt2" "Ly6c1" "Ly6a" "Sptbn1" "Gstm1"[121] "Selenop" "Jun" "Laptm4a" "Egr1" "Apoe" "Mt1"$`Grem1 fibroblast`[1] "Col6a3" "Mmp2" "Col6a2" "Grem1" "Fbn1" "Col5a1" "Tnc" "Nrep" "Col6a1" "Pcolce" "Col5a2"[12] "Col1a1" "Serpine2" "Col1a2" "Timp1" "Aebp1" "Bgn" "Col3a1" "Fgfr1" "Serpinh1" "Rbp1" "Adh1"[23] "Nbl1" "Cald1" "Gpx3" "Sod3" "Mfap4" "Olfml3" "Cp" "Mxra8" "Serping1" "Dcn" "Hmcn1"[34] "Colec12" "Mylk" "Sparcl1" "Sparc" "Rarres2" "Mfap5" "Rcn3" "Nfib" "Col4a1" "Fermt2" "Ltbp4"[45] "Ppic" "Igfbp7" "Ccn1" "Fmo2" "Tpm1" "Fstl1" "Timp3" "Gng11" "Nedd4" "Mgp" "Saa3"[56] "Mmp14" "Pmepa1" "Cfh" "Igfbp4" "Nupr1" "Plpp3" "Tmem176b" "Pbx1" "Tmem176a" "Tcf4" "Inmt"[67] "Zbtb20" "Id3" "Fn1" "Gsn"$Myofibroblast[1] "Tagln" "Myl9" "Tpm2" "Myh11" "Cald1" "Mustn1" "Sod3" "Ppp1r14a" "Mylk" "Actc1" "Des"[12] "Cnn1" "Fxyd1" "Ltbp4" "Cpe" "Map1b" "Lmod1" "Itih4" "Eln" "Serpine2" "Gucy1a1" "Actg2"[23] "Itga8" "Prss23" "Gucy1b1" "Postn" "Smtn" "Ndrg2" "Col18a1" "Synpo2" "Cdh13" "Hspg2" "Lamb2"[34] "Notch3" "Pdgfrb" "Ccn2" "Cox4i2" "Npnt" "Lmcd1" "Ccl19" "Sh3bgr" "Bgn" "Lhfp" "Rarres2"[45] "Tnxb" "Hspb6" "Sparcl1" "Col6a2" "Nr2f2" "Gm13889" "Snhg18" "Pde5a" "Aebp1" "Fhl1" "Cryab"[56] "Acta2" "Tinagl1" "Col1a2" "Mxra8" "Tgfb1i1" "Cavin3" "Col3a1" "Col6a1" "Timp3" "Igfbp5" "Fstl1"[67] "Ogn" "Serpinh1" "Selenom" "Tm4sf1" "Fermt2" "Col1a1" "Rcn3" "Pls3" "Sparc" "Fblim1" "Tpm1"[78] "Igfbp7" "Rbp1" "Mfge8" "Hspb1" "Nedd4" "Clu" "Cavin1" "Pam" "Ecm1" "Cnn3" "Bcam"[89] "Nfia" "Itga1" "Csrp2" "Col4a1" "Ppp1r12b" "Ccn1" "Tns1" "Ckb" "Crip2" "Csrp1" "Thra"[100] "Cd81" "Mgp" "Klf9" "Mcam" "Gpx3" "Rbpms" "Id3" "Lpp" "Tsc22d1" "Filip1l" "Dst"[111] "Nenf" "Zbtb20" "Gsn" "Epas1" "Dstn" "Slc25a4" "Selenow" "Actn1" "Itgb1" "Gstm1" "Flna"$Monocyte[1] "Plac8" "Ms4a6c" "Ifitm3" "S100a4" "Csf1r" "Pld4" "Cybb" "Ifitm6" "Pou2f2" "Napsa" "Apoe" "Ccr2" "Ace"$AM3[1] "Gpnmb" "Spp1" "Ccl2" "Fabp5" "Inhba" "Esd" "Cd63" "Cd68" "Ctsb" "Lgals3" "Cstb"[12] "Capg" "Rnh1" "Ctss" "Anxa4" "Rgcc" "Vat1" "Ctsz" "Emp1" "Lgmn" "Grn" "Ftl1-ps1"[23] "Pgam1" "Anxa5" "Ccl7"$AM2[1] "S100a1" "Fabp4" "Atp6v0d2" "Ctsk" "Fabp5" "Lpl" "Mt1" "Gstm1" "Chil3"$`NK cell`[1] "Gzma" "Ccl5" "Nkg7" "AW112010" "Il2rb" "Irf8" "Prf1" "Klrb1c" "Gzmb" "Serpinb9"[11] "Ctsw" "Ms4a4b" "Klrd1" "Klre1" "Ncr1" "Klra4" "Klra8" "Serpinb6b" "Klrk1" "Sh2d2a"[21] "Ptprcap" "Sytl3" "Pik3r1" "Ifngr1" "Klra7" "Sept1" "Klra9" "Cma1"$AM1[1] "Lpl" "Plet1" "Chil3" "Ear2" "Abcg1" "Mrc1" "Ear1" "Ltc4s" "Tcf7l2" "Axl" "Il18"[12] "Hebp1" "Mt1" "Atp6v0d2" "Sirpa" "Nceh1" "Krt19" "Fabp1" "Krt79" "Klhdc4" "Plin2" "Lmo4"[23] "Ptms" "Aplp2" "Sgk1" "Cidec" "Cd164" "Dstn" "Sept9" "Ctsd" "S100a1" "Wfdc21" "Ptpn12"$`Dendritic cell`[1] "H2-Aa" "H2-Eb1" "H2-Ab1" "Cd74" "Ifi30" "S100a4" "Cxcl16"$`Endothelial cell-2`[1] "Emp2" "Igfbp7" "Ramp2" "Ly6c1" "Cldn5" "Kdr" "Cdh5" "Cyp4b1" "Adgrf5" "Epas1" "Tspan7"[12] "Icam2" "Calcrl" "Tmem100" "Pmp22" "Kitl" "Egfl7" "Bcam" "Crip2" "Pcdh1" "Cavin2" "Slc9a3r2"[23] "Pecam1" "Fmo1" "Acvrl1" "Slco2a1" "Sema3f" "Aqp1" "Ednrb" "Tbx3" "Esam" "Cd34" "Fibin"[34] "Eng" "Sept4" "Stmn2" "Clic5" "Scn7a" "Podxl" "Ecscr" "Clu" "Prx" "Tmem204" "Tspan18"[45] "Col4a1" "Plpp1" "Hspb1" "Tbx2" "Clec1a" "Tmcc2" "Rgs12" "Myct1" "Sema6a" "Col4a2" "Itga3"[56] "Foxf1" "Myzap" "Car4" "Hpgd" "Cav1" "Timp3" "Nhlrc2" "Nrp1" "Krt80" "Cav2" "Ly6a"[67] "Ace" "Bmpr2" "Thbd" "Cd36" "Id3" "Anxa3" "Ctla2a" "Ehd4" "S1pr1" "Ptp4a3" "Selenop"[78] "Sptbn1" "Tmem176a" "Fkbp1a" "Tspan13" "App" "Clic4"$`Cycling basal cell`[1] "Pclaf" "Stmn1" "Mki67" "Birc5"$`B cell`[1] "Cd79a" "Ly6d" "Ebf1" "Cd79b" "H2-Eb1" "Ighm" "H2-Aa" "Igkc" "H2-DMb2" "H2-Ab1" "Cd74" "Fcmr" "Iglc2"[14] "Ms4a1" "H2-Ob" "Ccr7" "Ighd" "Iglc3" "Mef2c" "Cd19" "H2-Oa" "Cd37" "Bank1" "Mzb1" "Fcrla" "Scd1"[27] "Cd55" "H2-DMa" "Ets1"$Neutrophil[1] "S100a9" "S100a8" "Retnlg" "Il1r2" "Hdc" "Csf3r" "G0s2" "Ifitm1"[9] "Lmnb1" "Acod1" "Il1b" "Slpi" "Pglyrp1" "Trem1" "Clec4d" "Mmp9"[17] "Mxd1" "C5ar1" "Cxcr2" "Grina" "Lrg1" "Hp" "Ifitm2" "Slc16a3"[25] "Lcn2" "Slc7a11" "Ccrl2" "H2-Q10" "Marcksl1" "Clec4e" "Cd300ld" "Cxcl2"[33] "Lst1" "Isg15" "Hcar2" "Ets2" "Pla2g7" "Gadd45b" "Tnfaip2" "Msrb1"[41] "Cd14" "Stfa2l1" "Ccr1" "Srgn" "Arg2" "Wfdc17" "Ptgs2" "Sorl1"[49] "Sirpb1b" "Nfkbiz" "Slfn1" "Snx20" "Alox5ap" "2310001H17Rik"$`T cell`[1] "Cd3d" "Cd3e" "Cd3g" "Ms4a4b" "Trbc2" "Lat" "Il7r" "Thy1" "Ets1" "Gramd3" "Cd28" "H2-Q7"$IM[1] "C1qb" "C1qa" "C1qc" "Ms4a7" "Pf4" "Mafb" "Trem2" "Apoe" "Lgmn" "Ccl7" "H2-Eb1" "H2-Ab1" "H2-Aa"$`AT2 cell-2`[1] "Wfdc2" "Cyp2f2" "Sftpa1" "Selenbp1" "Sftpb" "Ldhb" "Aldh1a1" "Sftpd"[9] "Gsta4" "Cxcl15" "Gsta3" "Gstm2" "B430010I23Rik" "Clic3" "Sec14l3" "Retnla"[17] "Cbr2" "Scgb3a2" "Dcxr" "Nupr1" "Scgb1a1" "Mgst1" "Cyb5a" "Hp"[25] "Prdx6"$`Igha+ AT2 cell`[1] "Igkv13-85" "Hbb-bs" "Ighv1-64" "Scgb3a1" "Jchain" "Hbb-bt" "Spp1" "Mgp" "Sftpc" "Scgb3a2"[11] "Igha" "Scgb1a1"$`Endothelial cell-1`[1] "Ramp2" "Epas1" "Tspan7" "Ptprb" "Adgrf5" "Cdh5" "Hpgd" "Egfl7" "Ly6c1" "Calcrl" "Gpihbp1"[12] "Cldn5" "Plvap" "Pltp" "Tmem100" "Ly6a" "Cav1" "Cavin2" "Tm4sf1" "Pecam1" "Eng" "Slco2a1"[23] "Ctla2a" "Sema3c" "Cyyr1" "Crip2" "Clec14a" "Slc9a3r2" "Ace" "Sptbn1" "Acvrl1" "Cd36" "Ehd4"[34] "Cd93" "Aqp1" "Bmpr2" "Itga1" "Id3" "Sparc" "Esam" "BC028528" "Icam2" "Thbd" "S100a16"[45] "Col4a1" "Plpp1" "Fmo1" "Cav2" "Cavin1" "Cyp4b1" "Clec1a" "Tek" "Adgrl4" "Scn7a" "Flt1"[56] "Podxl" "Cxcl12" "Kit" "Tie1" "Foxf1" "Acer2" "Ece1" "Ecscr" "Cd200" "Gng11" "Nfib"[67] "Slc43a3" "Tjp1" "Col4a2" "Heg1" "Myzap" "Glp1r" "Lyve1" "Itga6" "Pakap.1" "Fermt2" "Rasip1"[78] "Pcdh17" "Jup" "Kdr" "Arhgef12" "Hmcn1" "Bmp6" "Clic5" "Dlc1" "Tcf4" "S1pr1" "Ccdc85b"[89] "Clec2d" "Jun" "Fkbp1a" "Clic4" "Tspan13" "Selenop"$`AT1 cell`[1] "Ager" "Cldn18" "Krt7" "Clic3" "Myh14" "Akap5" "Cyp2b10" "Rtkn2" "Mal2" "Igfbp2" "Aqp5"[12] "Lmo7" "Ndnf" "Myo1b" "Tspan8" "Gprc5a" "Limch1" "Cryab" "Clic5" "Emp2" "Sec14l3" "Bcam"[23] "Sparc" "Cystm1" "Igfbp7" "Crip2" "Timp3" "Mettl7a1" "Hopx" "Scd2" "Pmp22" "Spint2" "Ndst1"[34] "Vegfa"$`Clara cell`[1] "Scgb3a1" "Scgb3a2" "Bpifa1" "Bpifb1" "Reg3g" "Wfdc2" "Cyp2f2" "Mgp" "Scgb1a1" "Cbr2" "Sftpc" "Hbb-bs"$`Ciliated cell`[1] "Dynlrb2" "Hbb-bs" "Hbb-bt" "Cbr2"$`AT2 cell-1`[1] "Sftpc" "Cbr2" "Sftpa1" "Wfdc2" "Sftpb" "Sftpd" "Retnla" "Cyp2f2" "Scgb3a2" "Scgb1a1" "Chil1"$`Ig-producing B cell`[1] "Iglv1" "Mzb1" "Iglc1" "Derl3" "Eaf2" "Iglc3" "Jchain" "Iglc2" "Igha" "Igkc"[11] "Txndc5" "Cd79a" "Edem2" "Tent5c" "Creld2" "Prdx4" "Ly6d" "Lman1" "Trp53inp1" "Sdf2l1"[21] "Pdia4" "Edem1" "Fkbp2" "Xbp1" "Sec11c" "Manf" "Ssr4" "Hsp90b1" "Spcs1" "Igkv1-110"[31] "Ly6a"
制备好上述数据之后,运行下面代码
N <- length(st.clusts)M <- length(sc.clusts)MIA.results <- matrix(0,nrow = M, ncol = N)row.names(MIA.results) <- sc.clustscolnames(MIA.results) <- st.clustshead(MIA.results)# Gene universegene.universe <- length(rownames(cluster_gene_stat))# Loop over ST clustersfor (i in 1:N) {# Then loop over SC clusters# i=1for (j in 1:M) {genes1 <- st.marker.list[[st.clusts[i]]]genes2 <- sc.marker.list[[sc.clusts[j]]]# HypergeometricA <- length(intersect(genes1,genes2))B <- length(genes1)C <- length(genes2)enr <- -log10(phyper(A, B, gene.universe-B, C, lower.tail = FALSE))dep <- -log10(1-phyper(A, B, gene.universe-B, C, lower.tail = FALSE))if (enr < dep) {MIA.results[j,i] = -dep} else {MIA.results[j,i] = enr}}}# Some results were -Inf...check why this is the case...MIA.results[is.infinite(MIA.results)] <- 0MIA.resultspheatmap::pheatmap(MIA.results,scale ='row' )MIA.results2=MIA.results %>%as.matrix()MIA.results2[ MIA.results >5 ] =5pheatmap::pheatmap(MIA.results2,scale ='row' )# Visualize as heatmaplibrary(reshape2)library(ggplot2)library(dplyr)library(Seurat)heatmap_df <- data.frame('Cell types' = melt(MIA.results)[,1],'Tissue regions' = melt(MIA.results)[,2],enrichment = melt(MIA.results)[,3])range(heatmap_df$enrichment)head(heatmap_df)boxplot(heatmap_df$enrichment)#根据具体数值,设置最大值和最小值heatmap_df[abs(heatmap_df$enrichment) >7,]$enrichment=7ggplot(data = heatmap_df, aes(x = Tissue.regions, y = Cell.types, fill = enrichment)) +geom_tile() +scale_fill_gradient2(low = "navyblue", high = "red", mid = "white",midpoint = 0, limit = c(-7,7), space = "Lab",name="Enrichment \n -log10(p)") +ylim(heatmap_df$Cell.types %>% levels() %>% sort() %>% rev())+theme_minimal()+RotatedAxis()library(reshape2)library(ggplot2)library(dplyr)library(Seurat)library(scales)heatmap_df <- data.frame('Cell types' = melt(MIA.results)[,1],'Tissue regions' = melt(MIA.results)[,2],enrichment = melt(MIA.results)[,3])head(heatmap_df)ggplot(data = heatmap_df, aes(x = Tissue.regions, y = Cell.types, fill = enrichment)) +geom_tile() +scale_fill_gradient2(low = "blue", high = "red",mid = 'white',midpoint = 0, limit = c(-1,1), space = "Lab",oob=squish,name="Enrichment \n -log10(p)") +ylim(heatmap_df$Cell.types %>% levels() %>% sort() %>% rev())+theme_minimal()+RotatedAxis()
MIA.results,就是最终的结果。但是这里的热图具体怎么画,其实就是各显神通了
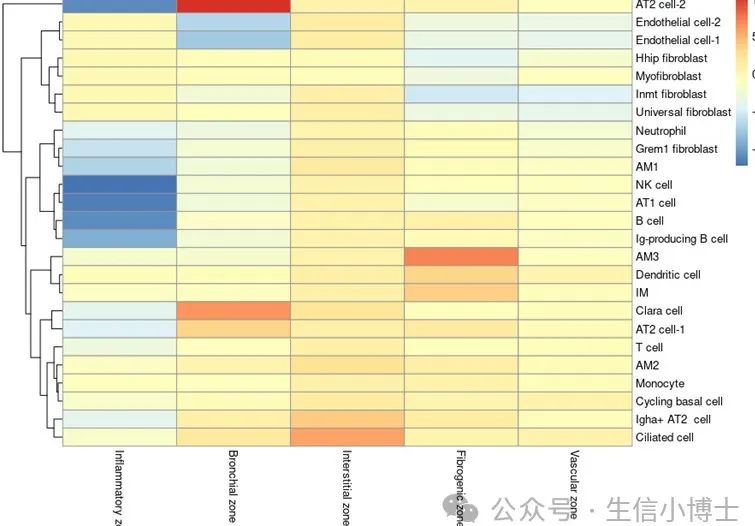
-
最后附上完整版本代码
#request 2.libPaths(c( "/home/data/t040413/R/x86_64-pc-linux-gnu-library/4.2","/home/data/t040413/R/yll/usr/local/lib/R/site-library","/home/data/refdir/Rlib/", "/usr/local/lib/R/library"))setwd('/home/data/t040413/silicosis/spatial/')library(Seurat)library(dplyr)#d.all=readRDS("~/silicosis/spatial/integrated_slides/integrated_slides.rds")load("/home/data/t040413/silicosis/spatial_transcriptomics/silicosis_ST_harmony_SCT_r0.5.rds")dim(d.all)DefaultAssay(d.all)="Spatial"#visium_slides=SplitObject(object = d.all,split.by = "stim")names(d.all);dim(d.all)d.all@meta.data %>%head()head(colnames(d.all))#给d.all 添加meta信息------adata_obs=read.csv("~/silicosis/spatial/adata_obs.csv")head(adata_obs)mymeta= paste0(d.all@meta.data$orig.ident,"_",colnames(d.all)) %>% gsub("-.*","",.) # %>% head()head(mymeta)tail(mymeta)#掉-及其之后内容adata_obs$col= adata_obs$spot_id %>% gsub("-.*","",.) # %>% head()head(adata_obs)rownames(adata_obs)=adata_obs$coladata_obs=adata_obs[mymeta,]head(adata_obs)identical(mymeta,adata_obs$col)d.all=AddMetaData(d.all,metadata = adata_obs)DefaultAssay(d.all)='Spatial'DimPlot(d.all,group.by = 'stim')DimPlot(d.all,label = TRUE)dev.off()####st-----cluster_gene_stat=FindAllMarkers(d.all,only.pos = TRUE,logfc.threshold = 0.5)head(cluster_gene_stat)table(cluster_gene_stat$cluster)# Initialize a dataframe for us to store values in:st.clusts=paste0("cluster",seq(0,8,1))head(cluster_gene_stat)st.marker.list=split(cluster_gene_stat,f = cluster_gene_stat$cluster)st.marker.list = lapply(st.marker.list,rownames)head(st.marker.list)#st.clusts=lapply(st.marker.list,rownames)st.clustsst.marker.list###sc----cellType_gene_stat =readRDS("~/silicosis/spatial2/marker_list_silicosis_nogrem1.rds")head(cellType_gene_stat)table(cellType_gene_stat$cluster)cellType_marker = list()for(i in unique(cellType_gene_stat$cluster)){ #按照标准 获得20个细胞类型的marker基因 列表cellType_marker[[i]] = cellType_gene_stat[cellType_gene_stat$cluster==i & cellType_gene_stat$p_val_adj<0.1 &cellType_gene_stat$avg_log2FC>0.7, "gene"]}cellType_markerhead(cellType_marker)names(cellType_marker)sc.marker.list=cellType_markersc.clusts=names(cellType_marker)sc.clustscellType_markerst.clustsst.marker.listN <- length(st.clusts)M <- length(sc.clusts)MIA.results <- matrix(0,nrow = M, ncol = N)row.names(MIA.results) <- sc.clustscolnames(MIA.results) <- st.clustshead(MIA.results)# Gene universegene.universe <- length(rownames(cluster_gene_stat))# Loop over ST clustersfor (i in 1:N) {# Then loop over SC clusters# i=1for (j in 1:M) {genes1 <- st.marker.list[[st.clusts[i]]]genes2 <- sc.marker.list[[sc.clusts[j]]]# HypergeometricA <- length(intersect(genes1,genes2))B <- length(genes1)C <- length(genes2)enr <- -log10(phyper(A, B, gene.universe-B, C, lower.tail = FALSE))dep <- -log10(1-phyper(A, B, gene.universe-B, C, lower.tail = FALSE))if (enr < dep) {MIA.results[j,i] = -dep} else {MIA.results[j,i] = enr}}}# Some results were -Inf...check why this is the case...MIA.results[is.infinite(MIA.results)] <- 0MIA.resultspheatmap::pheatmap(MIA.results,scale ='row' )MIA.results2=MIA.results %>%as.matrix()MIA.results2[ MIA.results >5 ] =5pheatmap::pheatmap(MIA.results2,scale ='row' )# Visualize as heatmaplibrary(reshape2)library(ggplot2)library(dplyr)library(Seurat)heatmap_df <- data.frame('Cell types' = melt(MIA.results)[,1],'Tissue regions' = melt(MIA.results)[,2],enrichment = melt(MIA.results)[,3])range(heatmap_df$enrichment)head(heatmap_df)boxplot(heatmap_df$enrichment)heatmap_df[abs(heatmap_df$enrichment) >7,]$enrichment=7ggplot(data = heatmap_df, aes(x = Tissue.regions, y = Cell.types, fill = enrichment)) +geom_tile() +scale_fill_gradient2(low = "navyblue", high = "red", mid = "white",midpoint = 0, limit = c(-7,7), space = "Lab",name="Enrichment \n -log10(p)") +ylim(heatmap_df$Cell.types %>% levels() %>% sort() %>% rev())+theme_minimal()+RotatedAxis()library(reshape2)library(ggplot2)library(dplyr)library(Seurat)library(scales)heatmap_df <- data.frame('Cell types' = melt(MIA.results)[,1],'Tissue regions' = melt(MIA.results)[,2],enrichment = melt(MIA.results)[,3])head(heatmap_df)ggplot(data = heatmap_df, aes(x = Tissue.regions, y = Cell.types, fill = enrichment)) +geom_tile() +scale_fill_gradient2(low = "blue", high = "red",mid = 'white',midpoint = 0, limit = c(-1,1), space = "Lab",oob=squish,name="Enrichment \n -log10(p)") +ylim(heatmap_df$Cell.types %>% levels() %>% sort() %>% rev())+theme_minimal()+RotatedAxis()
------------------------------------------------------------------------
最后,欢迎读者给出建设性意见,如有错误,欢迎批评指正。
——生信小博士